Parameterized level set optimization of a y-branch#
Note: the cost of running the entire notebook is higher than 1 FlexCredit.
This notebook demonstrates how to set up and run a parameterized level set-based optimization of a Y-branch. In this approach, we use autograd to generate a level set surface \(\phi(\rho)\) given a set of control knots \(\rho\). The permittivity distribution is then obtained implicitly from the zero level set isocontour. Details about the level set method can be found here. Minimum gap and curvature penalty terms are introduced in
the optimization to control the minimum feature size, hence improving device fabrication. In addition, we show how to tailor the initial level set function to a starting geometry, which is helpful to further optimize a device obtained by conventional design.
You can also find some interesting adjoint functionalities for shape optimization in Inverse design optimization of a waveguide taper and Adjoint-based shape optimization of a waveguide bend. If you are new to the finite-difference time-domain (FDTD) method, we highly recommend going through our FDTD101 tutorials. FDTD simulations can diverge due to various reasons. If you run into any simulation divergence issues, please follow the steps outlined in our troubleshooting guide to resolve it.
Let’s start by importing the Python libraries used throughout this notebook.
[1]:
# Standard python imports.
import pickle
from typing import List
# Import autograd to be able to use automatic differentiation.
import autograd.numpy as anp
import gdstk
import matplotlib.pylab as plt
import numpy as np
import optax
# Import regular tidy3d.
import tidy3d as td
import tidy3d.web as web
from autograd import grad
from autograd.tracer import getval
from tidy3d.plugins.autograd import value_and_grad
plt.rcParams["font.size"] = "12"
Y-branch Inverse Design Configuration#
The y-branch splits the power from an input waveguide into two other output waveguides. Here, we are considering a gap of 0.3 \(\mu m\) between the output waveguides for illustration purposes. However, when considering the design of a practical device, this value can be smaller. S-bends are included to keep the output waveguides apart from each other to prevent mode coupling.
Next, you can set the y-branch geometry and the inverse design parameters.
[2]:
# Geometric parameters.
y_width = 1.7 # Y-branch maximum width (um).
y_length = 1.7 # Y-branch maximum length (um).
w_thick = 0.22 # Waveguide thickness (um).
w_width = 0.5 # Waveguide width (um).
w_length = 1.0 # Input output waveguide length (um).
w_gap = 0.3 # Gap between the output waveguides (um).
bend_length = 3 # Output waveguide bend length (um).
bend_offset = 0.5 # Offset between output bends (um).
# Material.
nSi = 3.48 # Silicon refractive index.
# Inverse design set up parameters.
grid_size = 0.016 # Simulation grid size on design region (um).
ls_grid_size = 0.004 # Discretization size of the level set function (um).
ls_down_sample = (
20 # The spacing between the level set control knots is given by ls_grid_size*ls_down_sample.
)
fom_name_1 = "fom_field1" # Name of the monitor used to compute the objective function.
min_feature_size = 0.14 # Minimum fabrication feature size (um).
gap_par = 1.0 # Parameter to minimum gap fabrication constraint.
curve_par = 1.5 # Parameter of minimum curvature fabrication constraint.
# Optimizer parameters.
iterations = 100 # Maximum number of iterations in optimization.
learning_rate = 0.03
# Simulation wavelength.
wl = 1.55 # Central simulation wavelength (um).
bw = 0.06 # Simulation bandwidth (um).
n_wl = 61 # Number of wavelength points within the bandwidth.
From the parameters defined before, a lot of variables are computed and used to set up the optimization.
[3]:
# Minimum and maximum values for the permittivities.
eps_max = nSi**2
eps_min = 1.0
# Material definition.
mat_si = td.Medium(permittivity=eps_max) # Waveguide material.
# Wavelengths and frequencies.
wl_max = wl + bw / 2
wl_min = wl - bw / 2
wl_range = np.linspace(wl_min, wl_max, n_wl)
freq = td.C_0 / wl
freqs = td.C_0 / wl_range
freqw = 0.5 * (freqs[0] - freqs[-1])
run_time = 8e-13
# Computational domain size.
pml_spacing = 0.6 * wl
size_x = 2 * w_length + y_length + bend_length
size_y = w_gap + 2 * (bend_offset + w_width + pml_spacing)
size_z = w_thick + 2 * pml_spacing
eff_inf = 10
# Source and monitor positions.
mon_w = 3 * w_width
mon_h = 5 * w_thick
# Separation between the level set control knots.
rho_size = ls_down_sample * ls_grid_size
# Number of points on the parameter grid (rho) and level set grid (phi)
nx_rho = int(y_length / rho_size) + 1
ny_rho = int(y_width / rho_size / 2) + 1
nx_phi = int(y_length / ls_grid_size) + 1
ny_phi = int(y_width / ls_grid_size / 2) + 1
npar = nx_rho * ny_rho
ny_rho *= 2
ny_phi *= 2
# Design region size
dr_size_x = (nx_phi - 1) * ls_grid_size
dr_size_y = (ny_phi - 1) * ls_grid_size
dr_center_x = -size_x / 2 + w_length + dr_size_x / 2
# xy coordinates of the parameter and level set grids.
x_rho = np.linspace(dr_center_x - dr_size_x / 2, dr_center_x + dr_size_x / 2, nx_rho)
x_phi = np.linspace(dr_center_x - dr_size_x / 2, dr_center_x + dr_size_x / 2, nx_phi)
y_rho = np.linspace(-dr_size_y / 2, dr_size_y / 2, ny_rho)
y_phi = np.linspace(-dr_size_y / 2, dr_size_y / 2, ny_phi)
Level Set Functions#
We are using autograd to implement a parameterized level set function so the gradients can be back-propagated from the permittivity distribution defined by the zero level set isocontour to the design variables (the control knots of the level set surface). The space between the control knots and the Gaussian function width obtains some control over the minimum feature size. Other types of radial basis functions can also be used in replacement of the Gaussian one employed here, such as
multiquadric splines or b-splines.
[4]:
class LevelSetInterp:
"""This class implements the level set surface using Gaussian radial basis functions."""
def __init__(
self,
x0: anp.ndarray = None,
y0: anp.ndarray = None,
z0: anp.ndarray = None,
sigma: float = None,
):
# Input data.
x, y = anp.meshgrid(y0, x0)
xy0 = anp.column_stack((x.reshape(-1), y.reshape(-1)))
self.xy0 = xy0
self.z0 = z0
self.sig = sigma
# Builds the level set interpolation model.
gauss_kernel = self.gaussian(self.xy0, self.xy0)
self.model = anp.dot(anp.linalg.inv(gauss_kernel), self.z0)
def gaussian(self, xyi, xyj):
dist = anp.sqrt(
(xyi[:, 1].reshape(-1, 1) - xyj[:, 1].reshape(1, -1)) ** 2
+ (xyi[:, 0].reshape(-1, 1) - xyj[:, 0].reshape(1, -1)) ** 2
)
return anp.exp(-(dist**2) / (2 * self.sig**2))
def get_ls(self, x1, y1):
xx, yy = anp.meshgrid(y1, x1)
xy1 = anp.column_stack((xx.reshape(-1), yy.reshape(-1)))
ls = self.gaussian(self.xy0, xy1).T @ self.model
return ls
# Function to plot the level set surface.
def plot_level_set(x0, y0, rho, x1, y1, phi):
y, x = np.meshgrid(y0, x0)
yy, xx = np.meshgrid(y1, x1)
fig = plt.figure(figsize=(12, 6), tight_layout=True)
ax1 = fig.add_subplot(1, 2, 1, projection="3d")
ax1.view_init(elev=45, azim=-45, roll=0)
ax1.plot_surface(xx, yy, phi, cmap="RdBu", alpha=0.8)
ax1.contourf(
xx,
yy,
phi,
levels=[np.amin(phi), 0],
zdir="z",
offset=0,
colors=["k", "w"],
alpha=0.5,
)
ax1.contour3D(xx, yy, phi, 1, cmap="binary", linewidths=[2])
ax1.scatter(x, y, rho, color="black", linewidth=1.0)
ax1.set_title("Level set surface")
ax1.set_xlabel(r"x ($\mu m$)")
ax1.set_ylabel(r"y ($\mu m$)")
ax1.xaxis.pane.fill = False
ax1.yaxis.pane.fill = False
ax1.zaxis.pane.fill = False
ax1.xaxis.pane.set_edgecolor("w")
ax1.yaxis.pane.set_edgecolor("w")
ax1.zaxis.pane.set_edgecolor("w")
ax2 = fig.add_subplot(1, 2, 2)
ax2.contourf(xx, yy, phi, levels=[0, np.amax(phi)], colors=[[0, 0, 0]])
ax2.set_title("Zero level set contour")
ax2.set_xlabel(r"x ($\mu m$)")
ax2.set_ylabel(r"y ($\mu m$)")
ax2.set_aspect("equal")
plt.show()
To map the permittivities to the zero-level set contour and obtain continuous derivatives, we use a hyperbolic tangent function as an approximation to a Heaviside function. Other smooth functions, such as sigmoid and arctangent, can also be employed. As discussed here, the difference on computed interface using different functions will decrease when reducing the mesh size.
[5]:
def mirror_param(design_param):
param = anp.array(design_param).reshape((nx_rho, int(ny_rho / 2)))
try:
param_minus = param._value.copy()
except:
param_minus = param.copy()
return anp.concatenate((anp.fliplr(param_minus), param), axis=1).flatten()
def get_eps(design_param, sharpness=10.0, plot_levelset=False) -> np.ndarray:
"""Returns the permittivities defined by the zero level set isocontour"""
phi_model = LevelSetInterp(x0=x_rho, y0=y_rho, z0=design_param, sigma=rho_size)
phi = phi_model.get_ls(x1=x_phi, y1=y_phi)
# Calculates the permittivities from the level set surface
eps_phi = 0.5 * (anp.tanh(sharpness * phi) + 1)
eps = eps_min + (eps_max - eps_min) * eps_phi
eps = anp.maximum(eps, eps_min)
eps = anp.minimum(eps, eps_max)
# Reshapes the design parameters into a 2D matrix.
eps = anp.reshape(eps, (nx_phi, ny_phi))
# Plots the level set surface.
if plot_levelset:
rho = np.reshape(design_param, (nx_rho, ny_rho))
phi = np.reshape(phi, (nx_phi, ny_phi))
plot_level_set(x0=x_rho, y0=y_rho, rho=rho, x1=x_phi, y1=y_phi, phi=phi)
return eps
In the next function, the permittivity values are used to build a CustomMedium within the design region.
[6]:
def update_design(eps, unfold=False) -> List[td.Structure]:
# Reflects the structure about the x-axis.
eps_val = anp.array(eps).reshape((nx_phi, ny_phi, 1))
coords_x = [(dr_center_x - dr_size_x / 2) + ix * ls_grid_size for ix in range(nx_phi)]
if not unfold:
# Creation of a CustomMedium using the values of the design parameters.
coords_yp = [0 + iy * ls_grid_size for iy in range(int(ny_phi / 2))]
coords = dict(x=coords_x, y=coords_yp, z=[0])
eps_ag = td.SpatialDataArray(eps_val, coords=coords)
eps_medium = td.CustomMedium(permittivity=eps_ag)
box = td.Box(
center=(dr_center_x, dr_size_y / 4, 0),
size=(dr_size_x, dr_size_y / 2, w_thick),
)
structure = [td.Structure(geometry=box, medium=eps_medium)]
else:
# Creation of a CustomMedium using the values of the design parameters.
coords_y = [-dr_size_y / 2 + iy * ls_grid_size for iy in range(ny_phi)]
coords = dict(x=coords_x, y=coords_y, z=[0])
eps_ag = td.SpatialDataArray(eps_val, coords=coords)
eps_medium = td.CustomMedium(permittivity=eps_ag)
box = td.Box(center=(dr_center_x, 0, 0), size=(dr_size_x, dr_size_y, w_thick))
structure = [td.Structure(geometry=box, medium=eps_medium)]
return structure
Initial Structure#
We built an initial y-brach structure containing some holes and different gap sizes to demonstrate how the design evolves under fabrication constraints. We define this structure using a PolySlab object and then translate it into a permittivity grid of the same size as the one used to define the level set function. The holes are introduced in the polygon using the ClipOperation object.
[7]:
vertices = np.array(
[
(-size_x / 2 + w_length, w_width / 2),
(-size_x / 2 + w_length + 0.5, w_width / 2),
(-size_x / 2 + w_length + 0.75, w_gap / 2 + w_width),
(-size_x / 2 + w_length + dr_size_x, w_gap / 2 + w_width),
(-size_x / 2 + w_length + dr_size_x, w_gap / 2),
(-size_x / 2 + w_length + 2.5 * dr_size_x / 3, w_gap / 2),
(-size_x / 2 + w_length + 2.3 * dr_size_x / 3, w_gap / 6),
(-size_x / 2 + w_length + 1.8 * dr_size_x / 3, w_gap / 6),
(-size_x / 2 + w_length + 1.8 * dr_size_x / 3, -w_gap / 6),
(-size_x / 2 + w_length + 2.3 * dr_size_x / 3, -w_gap / 6),
(-size_x / 2 + w_length + 2.5 * dr_size_x / 3, -w_gap / 2),
(-size_x / 2 + w_length + dr_size_x, -w_gap / 2),
(-size_x / 2 + w_length + dr_size_x, -w_gap / 2 - w_width),
(-size_x / 2 + w_length + 0.75, -w_gap / 2 - w_width),
(-size_x / 2 + w_length + 0.5, -w_width / 2),
(-size_x / 2 + w_length, -w_width / 2),
]
)
y_poly = td.PolySlab(vertices=vertices, axis=2, slab_bounds=(-w_thick / 2, w_thick / 2))
y_hole1 = td.Cylinder(
center=(
-size_x / 2 + w_length + 1.7 * dr_size_x / 3,
w_gap / 2 + w_width / 1.75,
0,
),
radius=min_feature_size / 3,
length=w_thick,
axis=2,
)
y_hole2 = td.Cylinder(
center=(
-size_x / 2 + w_length + 1.7 * dr_size_x / 3,
-w_gap / 2 - w_width / 1.75,
0,
),
radius=min_feature_size / 3,
length=w_thick,
axis=2,
)
y_hole3 = td.Cylinder(
center=(
-size_x / 2 + w_length + 2.3 * dr_size_x / 3,
w_gap / 2 + w_width / 1.75,
0,
),
radius=min_feature_size / 1.5,
length=w_thick,
axis=2,
)
y_hole4 = td.Cylinder(
center=(
-size_x / 2 + w_length + 2.3 * dr_size_x / 3,
-w_gap / 2 - w_width / 1.75,
0,
),
radius=min_feature_size / 1.5,
length=w_thick,
axis=2,
)
init_design = td.ClipOperation(operation="difference", geometry_a=y_poly, geometry_b=y_hole1)
init_design = td.ClipOperation(operation="difference", geometry_a=init_design, geometry_b=y_hole2)
init_design = td.ClipOperation(operation="difference", geometry_a=init_design, geometry_b=y_hole3)
init_design = td.ClipOperation(operation="difference", geometry_a=init_design, geometry_b=y_hole4)
init_eps = init_design.inside_meshgrid(x=x_phi, y=y_phi, z=np.zeros(1))
init_eps = np.squeeze(init_eps) * eps_max
init_design.plot(z=0)
plt.show()
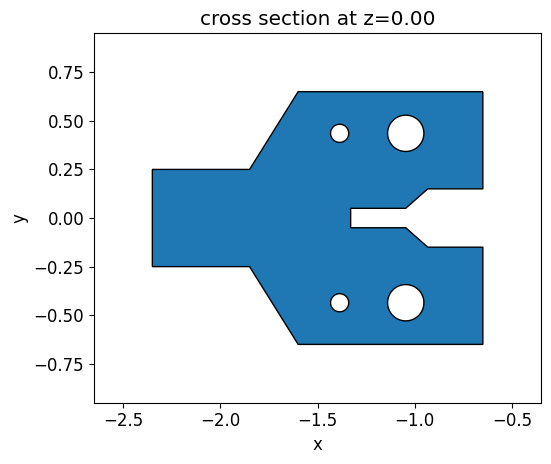
Then an objective function which compares the initial structure and the permittivity distribution generated by the level set zero contour is defined.
[8]:
# Figure of Merit (FOM) calculation.
def fom_eps(eps_ref: anp.ndarray, eps: anp.ndarray) -> float:
"""Calculate the L2 norm between eps_ref and eps."""
return anp.mean(anp.abs(eps_ref - eps) ** 2)
# Objective function to be passed to the optimization algorithm.
def obj_eps(design_param, eps_ref) -> float:
param = mirror_param(design_param)
eps = get_eps(param)
return fom_eps(eps_ref, eps)
# Function to calculate the objective function value and its
# gradient with respect to the design parameters.
obj_grad_eps = value_and_grad(obj_eps)
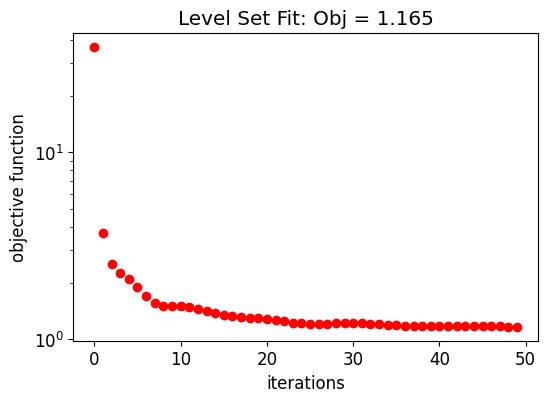
So, the initial control knots are obtained after fitting the initial structure using the level set function. This is accomplished by minimizing the L2 norm between the reference and the level set generated permittivities. The fitting is performed by the Adam optimizer from the Optax library.
[9]:
# Initialize adam optimizer with starting parameters.
start_par = np.zeros(npar)
optimizer = optax.adam(learning_rate=learning_rate * 10)
opt_state = optimizer.init(start_par)
# Store history
params_eps = np.copy(start_par)
obj_eps = []
for i in range(50):
# Compute gradient and current objective function value.
value, gradient = obj_grad_eps(params_eps, init_eps)
# outputs
print(f"Step = {i + 1}")
print(f"\tobj_eps = {value:.4e}")
print(f"\tgrad_norm = {np.linalg.norm(gradient):.4e}")
# Compute and apply updates to the optimizer based on gradient.
updates, opt_state = optimizer.update(gradient, opt_state, params_eps)
params_eps[:] = optax.apply_updates(params_eps, updates)
# Save history.
obj_eps.append(value)
# Gets the final parameters and the objective values history.
init_rho = np.copy(params_eps)
obj_vals_eps = np.array(obj_eps)
WARNING:2025-07-26 06:51:45,347:jax._src.xla_bridge:967: An NVIDIA GPU may be present on this machine, but a CUDA-enabled jaxlib is not installed. Falling back to cpu.
Step = 1
obj_eps = 3.6660e+01
grad_norm = 2.1337e+01
Step = 2
obj_eps = 3.8352e+00
grad_norm = 1.6936e+00
Step = 3
obj_eps = 2.5730e+00
grad_norm = 9.9867e-01
Step = 4
obj_eps = 2.2393e+00
grad_norm = 8.5924e-01
Step = 5
obj_eps = 2.0405e+00
grad_norm = 7.2283e-01
Step = 6
obj_eps = 1.7910e+00
grad_norm = 5.7901e-01
Step = 7
obj_eps = 1.5630e+00
grad_norm = 4.3837e-01
Step = 8
obj_eps = 1.4112e+00
grad_norm = 3.6683e-01
Step = 9
obj_eps = 1.3339e+00
grad_norm = 3.5290e-01
Step = 10
obj_eps = 1.3045e+00
grad_norm = 3.7025e-01
Step = 11
obj_eps = 1.2842e+00
grad_norm = 3.8085e-01
Step = 12
obj_eps = 1.2494e+00
grad_norm = 3.7093e-01
Step = 13
obj_eps = 1.1982e+00
grad_norm = 3.4044e-01
Step = 14
obj_eps = 1.1420e+00
grad_norm = 3.0237e-01
Step = 15
obj_eps = 1.0898e+00
grad_norm = 2.6239e-01
Step = 16
obj_eps = 1.0486e+00
grad_norm = 2.2298e-01
Step = 17
obj_eps = 1.0252e+00
grad_norm = 2.0291e-01
Step = 18
obj_eps = 1.0168e+00
grad_norm = 2.0348e-01
Step = 19
obj_eps = 1.0145e+00
grad_norm = 2.1126e-01
Step = 20
obj_eps = 1.0113e+00
grad_norm = 2.1521e-01
Step = 21
obj_eps = 1.0042e+00
grad_norm = 2.1250e-01
Step = 22
obj_eps = 9.9248e-01
grad_norm = 2.0371e-01
Step = 23
obj_eps = 9.7660e-01
grad_norm = 1.8586e-01
Step = 24
obj_eps = 9.5979e-01
grad_norm = 1.6358e-01
Step = 25
obj_eps = 9.4568e-01
grad_norm = 1.5022e-01
Step = 26
obj_eps = 9.3461e-01
grad_norm = 1.4719e-01
Step = 27
obj_eps = 9.2473e-01
grad_norm = 1.4254e-01
Step = 28
obj_eps = 9.1579e-01
grad_norm = 1.3176e-01
Step = 29
obj_eps = 9.0934e-01
grad_norm = 1.2223e-01
Step = 30
obj_eps = 9.0622e-01
grad_norm = 1.1917e-01
Step = 31
obj_eps = 9.0575e-01
grad_norm = 1.2234e-01
Step = 32
obj_eps = 9.0587e-01
grad_norm = 1.2684e-01
Step = 33
obj_eps = 9.0428e-01
grad_norm = 1.2751e-01
Step = 34
obj_eps = 8.9978e-01
grad_norm = 1.2027e-01
Step = 35
obj_eps = 8.9303e-01
grad_norm = 1.0461e-01
Step = 36
obj_eps = 8.8631e-01
grad_norm = 8.4470e-02
Step = 37
obj_eps = 8.8203e-01
grad_norm = 7.0781e-02
Step = 38
obj_eps = 8.8096e-01
grad_norm = 7.4915e-02
Step = 39
obj_eps = 8.8144e-01
grad_norm = 8.7472e-02
Step = 40
obj_eps = 8.8084e-01
grad_norm = 9.4230e-02
Step = 41
obj_eps = 8.7774e-01
grad_norm = 9.0045e-02
Step = 42
obj_eps = 8.7279e-01
grad_norm = 7.6119e-02
Step = 43
obj_eps = 8.6799e-01
grad_norm = 5.9258e-02
Step = 44
obj_eps = 8.6511e-01
grad_norm = 5.1586e-02
Step = 45
obj_eps = 8.6431e-01
grad_norm = 5.6978e-02
Step = 46
obj_eps = 8.6438e-01
grad_norm = 6.4243e-02
Step = 47
obj_eps = 8.6399e-01
grad_norm = 6.5572e-02
Step = 48
obj_eps = 8.6270e-01
grad_norm = 6.0463e-02
Step = 49
obj_eps = 8.6095e-01
grad_norm = 5.2884e-02
Step = 50
obj_eps = 8.5943e-01
grad_norm = 4.8189e-02
The following graph shows the evolution of the objective function along the initial structure fitting.
[10]:
fig, ax = plt.subplots(1, 1, figsize=(6, 4))
ax.plot(obj_vals_eps, "ro")
ax.set_xlabel("iterations")
ax.set_ylabel("objective function")
ax.set_title(f"Level Set Fit: Obj = {obj_vals_eps[-1]:.3f}")
ax.set_yscale("log")
plt.show()
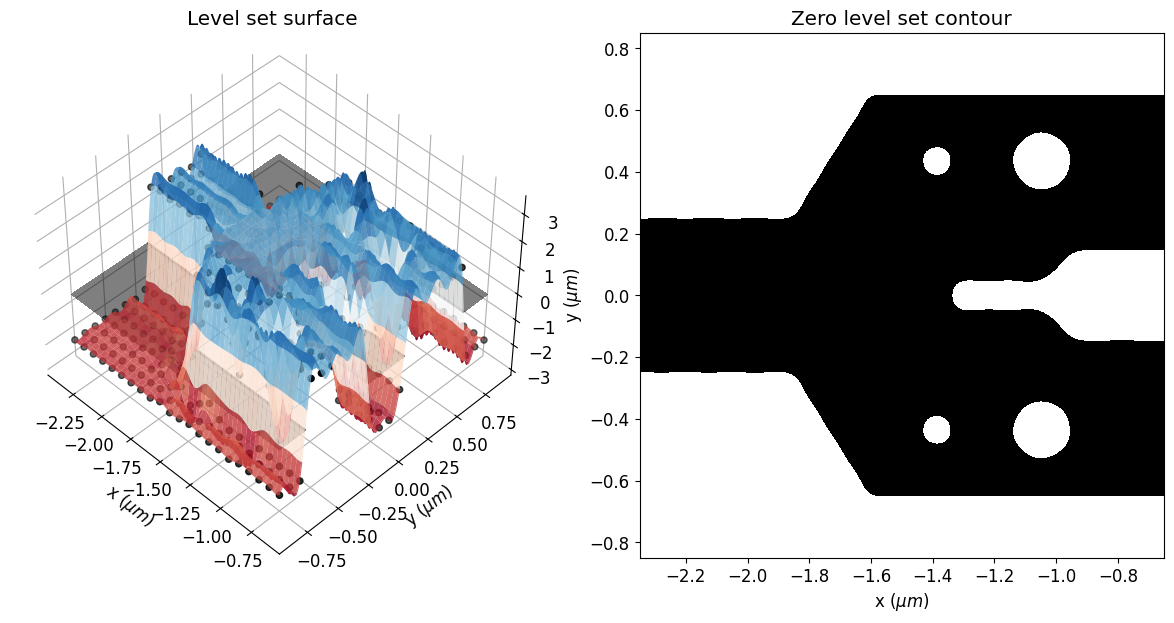
Here, one can see the initial parameters, which are the control knots defining the level set surface. The geometry of the structure will change as the zero isocontour evolves. The width of the Gaussian radial basis functions and the spacing of the control knots impact the accuracy and the smoothness of the initial zero-level set contour.
[11]:
eps_fit = get_eps(mirror_param(init_rho), plot_levelset=True)
Inverse Design Optimization Set Up#
Next, we will write a function to return the Simulation object. Note that we are using a MeshOverrideStructure to obtain a uniform mesh over the design region.
The elements that do not change along the optimization are defined first.
[12]:
# Input waveguide.
wg_input = td.Structure(
geometry=td.Box.from_bounds(
rmin=(-eff_inf, -w_width / 2, -w_thick / 2),
rmax=(-size_x / 2 + w_length + grid_size, w_width / 2, w_thick / 2),
),
medium=mat_si,
)
# Output bends.
x_start = (
-size_x / 2 + w_length + dr_size_x - grid_size
) # x-coordinate of the starting point of the waveguide bends.
x = np.linspace(x_start, x_start + bend_length, 100) # x-coordinates of the top edge vertices.
y = (
(x - x_start) * bend_offset / bend_length
- bend_offset * np.sin(2 * np.pi * (x - x_start) / bend_length) / (np.pi * 2)
+ (w_gap + w_width) / 2
) # y coordinates of the top edge vertices
# adding the last point to include the straight waveguide at the output
x = np.append(x, eff_inf)
y = np.append(y, y[-1])
# add path to the cell
cell = gdstk.Cell("bend")
cell.add(gdstk.FlexPath(x + 1j * y, w_width, layer=1, datatype=0)) # Top waveguide bend.
cell.add(gdstk.FlexPath(x - 1j * y, w_width, layer=1, datatype=0)) # Bottom waveguide bend.
# Define top waveguide bend structure.
wg_bend_top = td.Structure(
geometry=td.PolySlab.from_gds(
cell,
gds_layer=1,
axis=2,
slab_bounds=(-w_thick / 2, w_thick / 2),
)[1],
medium=mat_si,
)
# Define bottom waveguide bend structure.
wg_bend_bot = td.Structure(
geometry=td.PolySlab.from_gds(
cell,
gds_layer=1,
axis=2,
slab_bounds=(-w_thick / 2, w_thick / 2),
)[0],
medium=mat_si,
)
Monitors used to get simulation data.
[13]:
# Input mode source.
mode_spec = td.ModeSpec(num_modes=1, target_neff=nSi)
source = td.ModeSource(
center=(-size_x / 2 + 0.15 * wl, 0, 0),
size=(0, mon_w, mon_h),
source_time=td.GaussianPulse(freq0=freq, fwidth=freqw),
direction="+",
mode_spec=mode_spec,
mode_index=0,
)
# Monitor where we will compute the objective function from.
fom_monitor_1 = td.ModeMonitor(
center=[size_x / 2 - 0.25 * wl, (w_gap + w_width) / 2 + bend_offset, 0],
size=[0, mon_w, mon_h],
freqs=[freq],
mode_spec=mode_spec,
name=fom_name_1,
)
# Monitors used only to visualize the initial and final y-branch results.
# Field monitors to visualize the final fields.
field_xy = td.FieldMonitor(
size=(td.inf, td.inf, 0),
freqs=[freq],
name="field_xy",
)
# Monitor where we will compute the objective function from.
fom_final_1 = td.ModeMonitor(
center=[size_x / 2 - 0.25 * wl, (w_gap + w_width) / 2 + bend_offset, 0],
size=[0, mon_w, mon_h],
freqs=freqs,
mode_spec=mode_spec,
name="out_1",
)
And then the Simulation is built.
[14]:
def make_adjoint_sim(design_param, unfold=True) -> td.Simulation:
# Builds the design region from the design parameters.
eps = get_eps(design_param)
design_structure = update_design(eps, unfold=unfold)
# Creates a uniform mesh for the design region.
adjoint_dr_mesh = td.MeshOverrideStructure(
geometry=td.Box(center=(dr_center_x, 0, 0), size=(dr_size_x, dr_size_y, w_thick)),
dl=[grid_size, grid_size, grid_size],
enforce=True,
)
return td.Simulation(
size=[size_x, size_y, size_z],
center=[0, 0, 0],
grid_spec=td.GridSpec.auto(
wavelength=wl_max,
min_steps_per_wvl=15,
override_structures=[adjoint_dr_mesh],
),
symmetry=(0, -1, 1),
structures=[wg_input, wg_bend_top, wg_bend_bot] + design_structure,
sources=[source],
monitors=[fom_monitor_1],
run_time=run_time,
subpixel=True,
)
Let’s visualize the simulation setup and verify if all the elements are in their correct places. Differently from the density-based methods, we will start from a fully binarized structure.
[15]:
init_design = make_adjoint_sim(mirror_param(init_rho), unfold=True)
fig, ax1 = plt.subplots(1, 1, tight_layout=True, figsize=(8, 5))
init_design.plot_eps(z=0, ax=ax1)
plt.show()
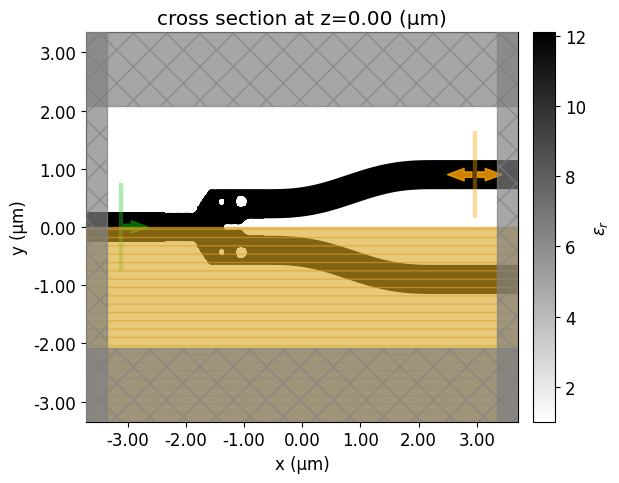
Now, we will run a simulation to see how this non-optimized y-branch performs.
[16]:
sim_init = init_design.copy(update=dict(monitors=(field_xy, fom_final_1)))
sim_data = web.run(sim_init, task_name="initial y-branch")
06:52:12 CEST Created task 'initial y-branch' with task_id 'fdve-b2696608-1a9c-4163-9bc3-406f5d847acb' and task_type 'FDTD'.
View task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-b2696608-1a 9c-4163-9bc3-406f5d847acb'.
Task folder: 'default'.
06:52:15 CEST Maximum FlexCredit cost: 0.025. Minimum cost depends on task execution details. Use 'web.real_cost(task_id)' to get the billed FlexCredit cost after a simulation run.
06:52:16 CEST status = queued
To cancel the simulation, use 'web.abort(task_id)' or 'web.delete(task_id)' or abort/delete the task in the web UI. Terminating the Python script will not stop the job running on the cloud.
06:52:25 CEST starting up solver
running solver
06:52:32 CEST early shutoff detected at 56%, exiting.
status = postprocess
06:52:34 CEST status = success
06:52:36 CEST View simulation result at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-b2696608-1a 9c-4163-9bc3-406f5d847acb'.
06:52:39 CEST loading simulation from simulation_data.hdf5
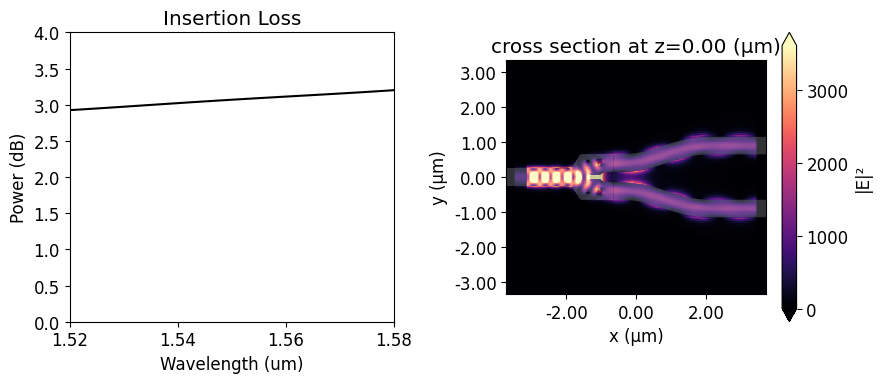
We will use the insertion loss (IL) to compare the device response before and after the optimization. Since we will use symmetry about the y-axis, the insertion loss is calculated as \(IL = -10 log(2P_{1}/P_{in})\), where \(P_{1}\) is the power coupled into the upper s-bend and \(P_{in}\) is the unit input power. The insertion loss of the non-optimized y-branch is above 3 dB at 1.55 \(\mu m\). From the field distribution image, we can realize that it happens because much of
the input power is reflected.
[17]:
coeffs_f = sim_data["out_1"].amps.sel(direction="+")
power_1 = np.abs(coeffs_f.sel(mode_index=0)) ** 2
power_1_db = -10 * np.log10(2 * power_1)
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(9, 4), tight_layout=True)
ax1.plot(wl_range, power_1_db, "-k")
ax1.set_xlabel("Wavelength (um)")
ax1.set_ylabel("Power (dB)")
ax1.set_ylim(0, 4)
ax1.set_xlim(wl - bw / 2, wl + bw / 2)
ax1.set_title("Insertion Loss")
sim_data.plot_field("field_xy", "E", "abs^2", z=0, ax=ax2)
plt.show()
Fabrication Constraints#
Fabrication constraints are introduced in the optimization as penalty terms to control the minimum gap (\(f_{g}\)) and radius of curvature (\(f_{c}\)) in the final design. Below, we use autograd to define the penalty terms following the formulation presented in D. Vercruysse, N. V. Sapra, L. Su, R. Trivedi, and J. Vučković, "Analytical level set fabrication constraints for inverse design," Scientific Reports 9, 8999 (2019). DOI:
10.1038/s41598-019-45026-0. The gap penalty function controls the minimum feature size by limiting the second derivative based on the value of the function at that point. The curvature constraint is only relevant at the device boundary, where \(\phi = 0\), so we apply the smoothed Heaviside function to the level set surface before calculating the derivatives.
[18]:
# Auxiliary function to calculate first and second order partial derivatives.
def ls_derivatives(phi, d_size):
SC = 1e-12
phi_1 = anp.array(anp.gradient(phi)) / d_size
phi_x = phi_1[0] + SC
phi_y = phi_1[1] + SC
phi_2x = anp.array(anp.gradient(phi_x)) / d_size
phi_2y = anp.array(anp.gradient(phi_y)) / d_size
phi_xx = phi_2x[0]
phi_xy = phi_2x[1]
phi_yy = phi_2y[1]
return phi_x, phi_y, phi_xx, phi_xy, phi_yy
# Minimum gap size fabrication constraint integrand calculation.
# The "beta" parameter relax the constraint near the zero plane.
def fab_penalty_ls_gap(params, beta=1, min_feature_size=min_feature_size, grid_size=ls_grid_size):
# Get the level set surface.
phi_model = LevelSetInterp(x0=x_rho, y0=y_rho, z0=params, sigma=rho_size)
phi = phi_model.get_ls(x1=x_phi, y1=y_phi)
phi = anp.reshape(phi, (nx_phi, ny_phi))
# Calculates their derivatives.
phi_x, phi_y, phi_xx, phi_xy, phi_yy = ls_derivatives(phi, grid_size)
# Calculates the gap penalty over the level set grid.
pi_d = np.pi / (1.3 * min_feature_size)
phi_v = anp.maximum(anp.power(phi_x**2 + phi_y**2, 0.5), anp.power(1e-32, 1 / 4))
phi_vv = (phi_x**2 * phi_xx + 2 * phi_x * phi_y * phi_xy + phi_y**2 * phi_yy) / phi_v**2
return (
anp.maximum((anp.abs(phi_vv) / (pi_d * anp.abs(phi) + beta * phi_v) - pi_d), 0)
* grid_size**2
)
# Minimum radius of curvature fabrication constraint integrand calculation.
# The "alpha" parameter controls its relative weight to the gap penalty.
# The "sharpness" parameter controls the smoothness of the surface near the zero-contour.
def fab_penalty_ls_curve(
params,
alpha=1,
sharpness=1,
min_feature_size=min_feature_size,
grid_size=ls_grid_size,
):
# Get the permittivity surface and calculates their derivatives.
eps = get_eps(params, sharpness=sharpness)
eps_x, eps_y, eps_xx, eps_xy, eps_yy = ls_derivatives(eps, grid_size)
# Calculates the curvature penalty over the permittivity grid.
pi_d = np.pi / (1.1 * min_feature_size)
eps_v = anp.maximum(anp.sqrt(eps_x**2 + eps_y**2), anp.power(1e-32, 1 / 6))
k = (eps_x**2 * eps_yy - 2 * eps_x * eps_y * eps_xy + eps_y**2 * eps_xx) / eps_v**3
curve_const = anp.abs(k * anp.arctan(eps_v / eps)) - pi_d
curve_const = anp.nan_to_num(curve_const)
return alpha * anp.maximum(curve_const, 0) * grid_size**2
# Gap and curvature fabrication constraints calculation.
# Penalty values are normalized by "norm_gap" and "norm_curve".
def fab_penalty_ls(
params,
beta=gap_par,
alpha=curve_par,
sharpness=4,
min_feature_size=min_feature_size,
grid_size=ls_grid_size,
norm_gap=1,
norm_curve=1,
):
# Get the gap penalty fabrication constraint value.
gap_penalty_int = fab_penalty_ls_gap(
params=params, beta=beta, min_feature_size=min_feature_size, grid_size=grid_size
)
gap_penalty_int = anp.nan_to_num(gap_penalty_int)
gap_penalty = anp.sum(gap_penalty_int) / norm_gap
# Get the curvature penalty fabrication constraint value.
curve_penalty_int = fab_penalty_ls_curve(
params=params,
alpha=alpha,
sharpness=sharpness,
min_feature_size=min_feature_size,
grid_size=grid_size,
)
curve_penalty_int = anp.nan_to_num(curve_penalty_int)
curve_penalty = anp.sum(curve_penalty_int) / norm_curve
return gap_penalty, curve_penalty
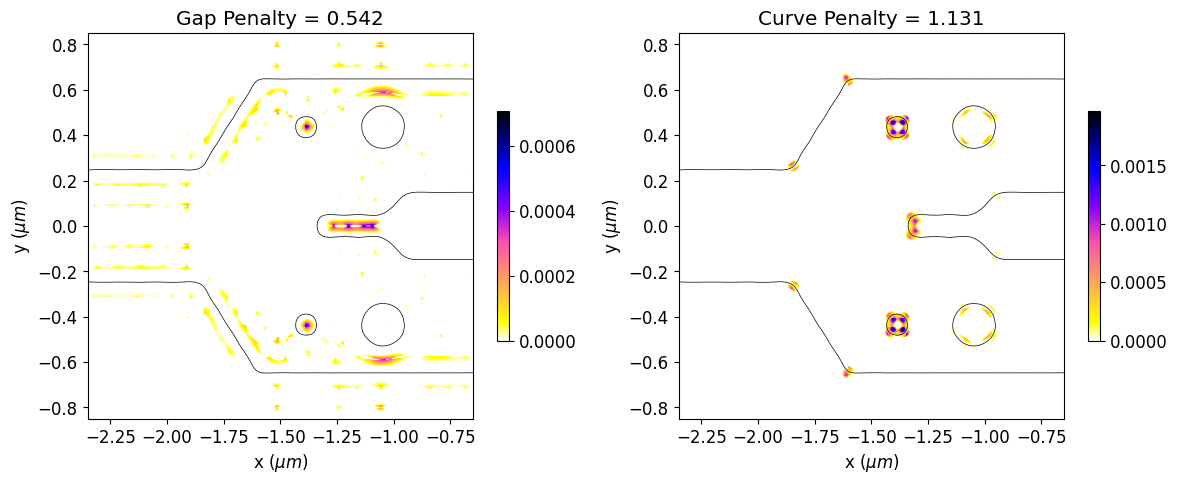
Now, we will calculate the initial penalty function values and observe the regions of the initial design that violate the constraints. The gap and curvature penalty functions are normalized by their initial values along the optimization to better balance the weights of device response and fabrication penalty within the objective function.
[19]:
# Initial values of gap and curvature fabrication constraints.
init_fab_gap, init_fab_curve = fab_penalty_ls(mirror_param(init_rho))
# Visualization of gap and curvature fabrication constraints values.
gap_penalty_int = fab_penalty_ls_gap(mirror_param(init_rho), beta=gap_par)
curve_penalty_int = fab_penalty_ls_curve(mirror_param(init_rho), alpha=curve_par, sharpness=4)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 8), tight_layout=True)
yy, xx = np.meshgrid(y_phi, x_phi)
im = ax1.imshow(
np.flipud(gap_penalty_int.T),
extent=[x_phi[0], x_phi[-1], y_phi[0], y_phi[-1]],
interpolation="none",
cmap="gnuplot2_r",
)
ax1.contour(xx, yy, eps_fit, [(eps_min + eps_max) / 2], colors="k", linewidths=0.5)
ax1.set_title(f"Gap Penalty = {init_fab_gap:.3f}")
ax1.set_xlabel(r"x ($\mu m$)")
ax1.set_ylabel(r"y ($\mu m$)")
fig.colorbar(im, ax=ax1, shrink=0.3)
im = ax2.imshow(
anp.flipud(curve_penalty_int.T),
extent=[x_phi[0], x_phi[-1], y_phi[0], y_phi[-1]],
interpolation="none",
cmap="gnuplot2_r",
)
ax2.contour(xx, yy, eps_fit, [(eps_min + eps_max) / 2], colors="k", linewidths=0.5)
ax2.set_title(f"Curve Penalty = {init_fab_curve:.3f}")
ax2.set_xlabel(r"x ($\mu m$)")
ax2.set_ylabel(r"y ($\mu m$)")
fig.colorbar(im, ax=ax2, shrink=0.3)
plt.show()
Running the Optimization#
The figure-of-merit used in the y-branch optimization is the power (\(P_{1, 2}\)) coupled into the fundamental transverse electric mode of the output waveguides. We will set mirror symmetry about the y-axis in the optimization, so we must include only \(P_{1}\) in the figure-of-merit. As we are using a minimization strategy, the coupled power and fabrication constraints are arranged within the objective function as \(|0.5 - P_{1}| + w_{f} \times (f_{g} + f_{c})\), where
\(w_{f}\) is the fabrication constraint weight, whereas \(f_{g}\) and \(f_{c}\) are the gap and curvature penalty values.
[20]:
# Figure of Merit (FOM) calculation.
def fom(sim_data: td.SimulationData) -> float:
"""Return the power at the mode index of interest."""
output_amps1 = sim_data[fom_name_1].amps
amp1 = output_amps1.sel(direction="+", f=freq, mode_index=0)
eta1 = anp.sum(anp.nan_to_num(anp.abs(amp1.values)) ** 2)
return anp.abs(0.5 - eta1), eta1
# Objective function to be passed to the optimization algorithm.
def obj(
design_param,
fab_const: float = 0.0,
norm_gap=1.0,
norm_curve=1.0,
verbose: bool = False,
) -> float:
param = mirror_param(design_param)
sim = make_adjoint_sim(param)
sim_data = web.run(sim, task_name="inv_des_ybranch", verbose=verbose)
fom_val, eta1 = fom(sim_data)
fab_gap, fab_curve = fab_penalty_ls(param, norm_gap=norm_gap, norm_curve=norm_curve)
J = fom_val + fab_const * (fab_gap + fab_curve)
aux_data = anp.array([sim_data, getval(eta1), getval(fab_gap), getval(fab_curve)])
return (J, aux_data)
# Function to calculate the objective function value and its
# gradient with respect to the design parameters.
obj_grad = value_and_grad(obj, has_aux=True)
Optimizer initialization
[21]:
# where to store history
history_fname = "./misc/y_branch_fab.pkl"
def save_history(history_dict: dict) -> None:
"""Convenience function to save the history to file."""
with open(history_fname, "wb") as file:
pickle.dump(history_dict, file)
def load_history() -> dict:
"""Convenience method to load the history from file."""
with open(history_fname, "rb") as file:
history_dict = pickle.load(file)
return history_dict
Before starting, we will look for data from a previous optimization.
[22]:
# Initialize adam optimizer with starting parameters.
optimizer = optax.adam(learning_rate=learning_rate * 8)
try:
history_dict = load_history()
opt_state = history_dict["opt_states"][-1]
params = history_dict["params"][-1]
num_iters_completed = len(history_dict["params"])
print("Loaded optimization checkpoint from file.")
print(f"Found {num_iters_completed} iterations previously completed out of {iterations} total.")
if num_iters_completed < iterations:
print("Will resume optimization.")
else:
print("Optimization completed, will return results.")
except (FileNotFoundError, IndexError):
params = np.array(init_rho)
opt_state = optimizer.init(params)
history_dict = dict(
values=[],
eta1=[],
penalty_gap=[],
penalty_curve=[],
params=[],
gradients=[],
opt_states=[opt_state],
data=[],
)
Now, we are ready to run the optimization!
[23]:
td.config.logging_level = "WARNING"
iter_done = len(history_dict["values"])
fab_const = 0.05
param_eps = anp.array(get_eps(mirror_param(anp.array(params))))
param_shape = param_eps.shape
param_eps = param_eps.flatten()
if iter_done < iterations:
for i in range(iter_done, iterations):
params = anp.array(params)
(value, gradient), aux = obj_grad(
params,
fab_const=fab_const,
norm_gap=init_fab_gap,
norm_curve=init_fab_curve,
)
sim_data_i, eta1, penalty_gap, penalty_curve = aux
gradient = np.nan_to_num(gradient)
gradient = np.array(gradient)
params = np.array(params)
# outputs
print(f"Step = {i + 1}")
print(f"\tobj_val = {value:.4e}")
print(f"\tgrad_norm = {np.linalg.norm(gradient):.4e}")
print(f"\teta1 = {eta1:.3f}")
print(f"\tpenalty gap = {penalty_gap:.3f}")
print(f"\tpenalty curve = {penalty_curve:.3f}")
# Compute and apply updates to the optimizer based on gradient.
updates, opt_state = optimizer.update(gradient, opt_state, params)
params = optax.apply_updates(params, updates)
# Save history.
history_dict["values"].append(value)
history_dict["eta1"].append(eta1)
history_dict["penalty_gap"].append(penalty_gap)
history_dict["penalty_curve"].append(penalty_curve)
history_dict["params"].append(params)
history_dict["gradients"].append(gradient)
history_dict["opt_states"].append(opt_state)
# history_dict["data"].append(sim_data_i) # Uncomment to store data, can create large files.
save_history(history_dict)
Step = 1
obj_val = 3.5564e-01
grad_norm = 5.3675e-02
eta1 = 0.244
penalty gap = 1.000
penalty curve = 1.000
Step = 2
obj_val = 3.0410e-01
grad_norm = 4.1640e-02
eta1 = 0.285
penalty gap = 0.531
penalty curve = 1.249
Step = 3
obj_val = 2.9423e-01
grad_norm = 4.3639e-02
eta1 = 0.324
penalty gap = 0.791
penalty curve = 1.573
Step = 4
obj_val = 2.6636e-01
grad_norm = 6.3644e-02
eta1 = 0.354
penalty gap = 0.868
penalty curve = 1.535
Step = 5
obj_val = 2.2304e-01
grad_norm = 9.3690e-02
eta1 = 0.401
penalty gap = 0.890
penalty curve = 1.581
Step = 6
obj_val = 1.9169e-01
grad_norm = 3.0088e-02
eta1 = 0.421
penalty gap = 0.731
penalty curve = 1.524
Step = 7
obj_val = 1.6666e-01
grad_norm = 4.1442e-02
eta1 = 0.431
penalty gap = 0.611
penalty curve = 1.344
Step = 8
obj_val = 1.3648e-01
grad_norm = 2.4416e-02
eta1 = 0.443
penalty gap = 0.498
penalty curve = 1.100
Step = 9
obj_val = 1.2933e-01
grad_norm = 1.6903e-02
eta1 = 0.448
penalty gap = 0.442
penalty curve = 1.107
Step = 10
obj_val = 1.2578e-01
grad_norm = 1.6739e-02
eta1 = 0.452
penalty gap = 0.424
penalty curve = 1.132
Step = 11
obj_val = 1.2318e-01
grad_norm = 1.7043e-02
eta1 = 0.454
penalty gap = 0.388
penalty curve = 1.156
Step = 12
obj_val = 1.2037e-01
grad_norm = 1.6390e-02
eta1 = 0.456
penalty gap = 0.362
penalty curve = 1.169
Step = 13
obj_val = 1.1856e-01
grad_norm = 1.7511e-02
eta1 = 0.456
penalty gap = 0.349
penalty curve = 1.147
Step = 14
obj_val = 1.1749e-01
grad_norm = 1.4882e-02
eta1 = 0.456
penalty gap = 0.368
penalty curve = 1.112
Step = 15
obj_val = 1.1523e-01
grad_norm = 1.4130e-02
eta1 = 0.458
penalty gap = 0.388
penalty curve = 1.081
Step = 16
obj_val = 1.1310e-01
grad_norm = 1.1825e-02
eta1 = 0.461
penalty gap = 0.418
penalty curve = 1.059
Step = 17
obj_val = 1.1141e-01
grad_norm = 1.0207e-02
eta1 = 0.463
penalty gap = 0.430
penalty curve = 1.055
Step = 18
obj_val = 1.1016e-01
grad_norm = 9.3851e-03
eta1 = 0.465
penalty gap = 0.426
penalty curve = 1.072
Step = 19
obj_val = 1.0877e-01
grad_norm = 8.3718e-03
eta1 = 0.467
penalty gap = 0.415
penalty curve = 1.102
Step = 20
obj_val = 1.0834e-01
grad_norm = 8.2702e-03
eta1 = 0.469
penalty gap = 0.409
penalty curve = 1.141
Step = 21
obj_val = 1.0747e-01
grad_norm = 8.0938e-03
eta1 = 0.471
penalty gap = 0.395
penalty curve = 1.174
Step = 22
obj_val = 1.0720e-01
grad_norm = 8.6709e-03
eta1 = 0.472
penalty gap = 0.388
penalty curve = 1.203
Step = 23
obj_val = 1.0724e-01
grad_norm = 8.5987e-03
eta1 = 0.473
penalty gap = 0.380
penalty curve = 1.223
Step = 24
obj_val = 1.0654e-01
grad_norm = 8.9436e-03
eta1 = 0.473
penalty gap = 0.368
penalty curve = 1.233
Step = 25
obj_val = 1.0543e-01
grad_norm = 9.6338e-03
eta1 = 0.475
penalty gap = 0.364
penalty curve = 1.235
Step = 26
obj_val = 1.0423e-01
grad_norm = 8.8936e-03
eta1 = 0.475
penalty gap = 0.359
penalty curve = 1.231
Step = 27
obj_val = 1.0284e-01
grad_norm = 7.6343e-03
eta1 = 0.476
penalty gap = 0.348
penalty curve = 1.229
Step = 28
obj_val = 1.0055e-01
grad_norm = 6.4258e-03
eta1 = 0.477
penalty gap = 0.322
penalty curve = 1.227
Step = 29
obj_val = 9.9215e-02
grad_norm = 6.8030e-03
eta1 = 0.478
penalty gap = 0.308
penalty curve = 1.227
Step = 30
obj_val = 9.8130e-02
grad_norm = 7.3612e-03
eta1 = 0.478
penalty gap = 0.300
penalty curve = 1.227
Step = 31
obj_val = 9.5986e-02
grad_norm = 6.5892e-03
eta1 = 0.480
penalty gap = 0.288
penalty curve = 1.225
Step = 32
obj_val = 9.4908e-02
grad_norm = 6.2832e-03
eta1 = 0.481
penalty gap = 0.285
penalty curve = 1.224
Step = 33
obj_val = 9.4727e-02
grad_norm = 7.4404e-03
eta1 = 0.481
penalty gap = 0.280
penalty curve = 1.233
Step = 34
obj_val = 9.3789e-02
grad_norm = 7.2913e-03
eta1 = 0.481
penalty gap = 0.276
penalty curve = 1.227
Step = 35
obj_val = 9.2355e-02
grad_norm = 6.2001e-03
eta1 = 0.482
penalty gap = 0.271
penalty curve = 1.218
Step = 36
obj_val = 9.0359e-02
grad_norm = 5.8173e-03
eta1 = 0.483
penalty gap = 0.265
penalty curve = 1.208
Step = 37
obj_val = 8.8227e-02
grad_norm = 5.8414e-03
eta1 = 0.485
penalty gap = 0.255
penalty curve = 1.201
Step = 38
obj_val = 8.6384e-02
grad_norm = 6.0266e-03
eta1 = 0.485
penalty gap = 0.246
penalty curve = 1.189
Step = 39
obj_val = 8.4375e-02
grad_norm = 5.6968e-03
eta1 = 0.486
penalty gap = 0.238
penalty curve = 1.172
Step = 40
obj_val = 8.2553e-02
grad_norm = 6.7060e-03
eta1 = 0.487
penalty gap = 0.232
penalty curve = 1.152
Step = 41
obj_val = 8.0284e-02
grad_norm = 5.1719e-03
eta1 = 0.487
penalty gap = 0.224
penalty curve = 1.126
Step = 42
obj_val = 7.8758e-02
grad_norm = 6.1781e-03
eta1 = 0.487
penalty gap = 0.222
penalty curve = 1.098
Step = 43
obj_val = 7.7261e-02
grad_norm = 6.1637e-03
eta1 = 0.487
penalty gap = 0.214
penalty curve = 1.070
Step = 44
obj_val = 7.5783e-02
grad_norm = 6.2459e-03
eta1 = 0.487
penalty gap = 0.212
penalty curve = 1.043
Step = 45
obj_val = 7.3889e-02
grad_norm = 5.0465e-03
eta1 = 0.487
penalty gap = 0.202
penalty curve = 1.021
Step = 46
obj_val = 7.2547e-02
grad_norm = 5.7386e-03
eta1 = 0.487
penalty gap = 0.199
penalty curve = 1.001
Step = 47
obj_val = 7.0999e-02
grad_norm = 6.0761e-03
eta1 = 0.488
penalty gap = 0.192
penalty curve = 0.978
Step = 48
obj_val = 6.9336e-02
grad_norm = 5.6293e-03
eta1 = 0.488
penalty gap = 0.192
penalty curve = 0.950
Step = 49
obj_val = 6.8035e-02
grad_norm = 4.4431e-03
eta1 = 0.488
penalty gap = 0.194
penalty curve = 0.922
Step = 50
obj_val = 6.6989e-02
grad_norm = 6.8384e-03
eta1 = 0.487
penalty gap = 0.191
penalty curve = 0.898
Step = 51
obj_val = 6.7029e-02
grad_norm = 1.0912e-02
eta1 = 0.487
penalty gap = 0.199
penalty curve = 0.878
Step = 52
obj_val = 6.5397e-02
grad_norm = 4.0047e-03
eta1 = 0.488
penalty gap = 0.199
penalty curve = 0.861
Step = 53
obj_val = 6.5150e-02
grad_norm = 7.3279e-03
eta1 = 0.487
penalty gap = 0.201
penalty curve = 0.845
Step = 54
obj_val = 6.4428e-02
grad_norm = 8.6795e-03
eta1 = 0.487
penalty gap = 0.198
penalty curve = 0.831
Step = 55
obj_val = 6.2940e-02
grad_norm = 5.3589e-03
eta1 = 0.487
penalty gap = 0.193
penalty curve = 0.812
Step = 56
obj_val = 6.1655e-02
grad_norm = 7.2568e-03
eta1 = 0.487
penalty gap = 0.187
penalty curve = 0.791
Step = 57
obj_val = 5.9985e-02
grad_norm = 4.4634e-03
eta1 = 0.488
penalty gap = 0.180
penalty curve = 0.772
Step = 58
obj_val = 5.8987e-02
grad_norm = 6.2809e-03
eta1 = 0.487
penalty gap = 0.172
penalty curve = 0.756
Step = 59
obj_val = 5.7932e-02
grad_norm = 6.6443e-03
eta1 = 0.487
penalty gap = 0.163
penalty curve = 0.744
Step = 60
obj_val = 5.7018e-02
grad_norm = 7.1717e-03
eta1 = 0.488
penalty gap = 0.159
penalty curve = 0.734
Step = 61
obj_val = 5.6096e-02
grad_norm = 7.1267e-03
eta1 = 0.488
penalty gap = 0.155
penalty curve = 0.723
Step = 62
obj_val = 5.5139e-02
grad_norm = 5.5468e-03
eta1 = 0.488
penalty gap = 0.154
penalty curve = 0.710
Step = 63
obj_val = 5.4309e-02
grad_norm = 5.3635e-03
eta1 = 0.488
penalty gap = 0.150
penalty curve = 0.693
Step = 64
obj_val = 5.3077e-02
grad_norm = 4.3196e-03
eta1 = 0.488
penalty gap = 0.145
penalty curve = 0.671
Step = 65
obj_val = 5.1967e-02
grad_norm = 5.7827e-03
eta1 = 0.488
penalty gap = 0.141
penalty curve = 0.652
Step = 66
obj_val = 5.0103e-02
grad_norm = 5.6972e-03
eta1 = 0.488
penalty gap = 0.135
penalty curve = 0.636
Step = 67
obj_val = 4.9044e-02
grad_norm = 4.3164e-03
eta1 = 0.488
penalty gap = 0.127
penalty curve = 0.621
Step = 68
obj_val = 4.7887e-02
grad_norm = 5.2754e-03
eta1 = 0.488
penalty gap = 0.117
penalty curve = 0.602
Step = 69
obj_val = 4.6736e-02
grad_norm = 4.5430e-03
eta1 = 0.488
penalty gap = 0.110
penalty curve = 0.581
Step = 70
obj_val = 4.5639e-02
grad_norm = 4.7783e-03
eta1 = 0.488
penalty gap = 0.103
penalty curve = 0.560
Step = 71
obj_val = 4.4642e-02
grad_norm = 5.8226e-03
eta1 = 0.487
penalty gap = 0.098
penalty curve = 0.543
Step = 72
obj_val = 4.4104e-02
grad_norm = 6.7811e-03
eta1 = 0.488
penalty gap = 0.094
penalty curve = 0.538
Step = 73
obj_val = 4.3684e-02
grad_norm = 1.3643e-02
eta1 = 0.487
penalty gap = 0.092
penalty curve = 0.519
Step = 74
obj_val = 4.3386e-02
grad_norm = 1.6012e-02
eta1 = 0.486
penalty gap = 0.089
penalty curve = 0.502
Step = 75
obj_val = 4.2177e-02
grad_norm = 7.5301e-03
eta1 = 0.487
penalty gap = 0.090
penalty curve = 0.489
Step = 76
obj_val = 4.3145e-02
grad_norm = 2.4119e-02
eta1 = 0.485
penalty gap = 0.088
penalty curve = 0.482
Step = 77
obj_val = 4.1368e-02
grad_norm = 1.0816e-02
eta1 = 0.486
penalty gap = 0.089
penalty curve = 0.467
Step = 78
obj_val = 4.1039e-02
grad_norm = 1.4039e-02
eta1 = 0.486
penalty gap = 0.088
penalty curve = 0.452
Step = 79
obj_val = 4.0349e-02
grad_norm = 1.2459e-02
eta1 = 0.486
penalty gap = 0.091
penalty curve = 0.436
Step = 80
obj_val = 3.9335e-02
grad_norm = 1.1502e-02
eta1 = 0.486
penalty gap = 0.088
penalty curve = 0.421
Step = 81
obj_val = 3.9219e-02
grad_norm = 1.3385e-02
eta1 = 0.486
penalty gap = 0.090
penalty curve = 0.412
Step = 82
obj_val = 3.8009e-02
grad_norm = 6.5264e-03
eta1 = 0.487
penalty gap = 0.082
penalty curve = 0.411
Step = 83
obj_val = 3.8244e-02
grad_norm = 1.4448e-02
eta1 = 0.486
penalty gap = 0.079
penalty curve = 0.409
Step = 84
obj_val = 3.6880e-02
grad_norm = 5.0679e-03
eta1 = 0.487
penalty gap = 0.074
penalty curve = 0.394
Step = 85
obj_val = 3.7210e-02
grad_norm = 1.4843e-02
eta1 = 0.486
penalty gap = 0.072
penalty curve = 0.387
Step = 86
obj_val = 3.6202e-02
grad_norm = 5.3322e-03
eta1 = 0.486
penalty gap = 0.070
penalty curve = 0.380
Step = 87
obj_val = 3.5769e-02
grad_norm = 9.8970e-03
eta1 = 0.486
penalty gap = 0.066
penalty curve = 0.372
Step = 88
obj_val = 3.5204e-02
grad_norm = 4.9286e-03
eta1 = 0.486
penalty gap = 0.061
penalty curve = 0.368
Step = 89
obj_val = 3.5098e-02
grad_norm = 1.0777e-02
eta1 = 0.486
penalty gap = 0.058
penalty curve = 0.362
Step = 90
obj_val = 3.4205e-02
grad_norm = 4.9976e-03
eta1 = 0.486
penalty gap = 0.060
penalty curve = 0.345
Step = 91
obj_val = 3.3863e-02
grad_norm = 1.1599e-02
eta1 = 0.485
penalty gap = 0.061
penalty curve = 0.322
Step = 92
obj_val = 3.3350e-02
grad_norm = 6.5625e-03
eta1 = 0.485
penalty gap = 0.058
penalty curve = 0.305
Step = 93
obj_val = 3.3549e-02
grad_norm = 1.2542e-02
eta1 = 0.484
penalty gap = 0.057
penalty curve = 0.296
Step = 94
obj_val = 3.3028e-02
grad_norm = 8.5376e-03
eta1 = 0.484
penalty gap = 0.058
penalty curve = 0.287
Step = 95
obj_val = 3.2332e-02
grad_norm = 8.3651e-03
eta1 = 0.485
penalty gap = 0.063
penalty curve = 0.278
Step = 96
obj_val = 3.1877e-02
grad_norm = 7.5040e-03
eta1 = 0.485
penalty gap = 0.062
penalty curve = 0.275
Step = 97
obj_val = 3.1225e-02
grad_norm = 5.6359e-03
eta1 = 0.485
penalty gap = 0.060
penalty curve = 0.267
Step = 98
obj_val = 3.0762e-02
grad_norm = 7.2721e-03
eta1 = 0.485
penalty gap = 0.056
penalty curve = 0.256
Step = 99
obj_val = 3.0489e-02
grad_norm = 1.5867e-02
eta1 = 0.485
penalty gap = 0.055
penalty curve = 0.250
Step = 100
obj_val = 3.0802e-02
grad_norm = 1.0551e-02
eta1 = 0.484
penalty gap = 0.051
penalty curve = 0.254
[24]:
obj_vals = np.array(history_dict["values"])
eta1_vals = np.array(history_dict["eta1"])
pen_gap_vals = np.array(history_dict["penalty_gap"])
pen_curve_vals = np.array(history_dict["penalty_curve"])
final_par = history_dict["params"][-1]
Optimization Results#
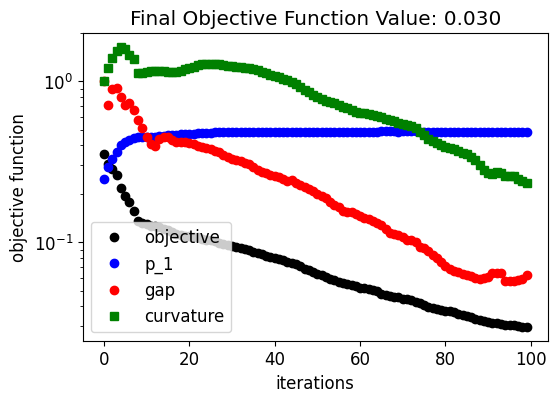
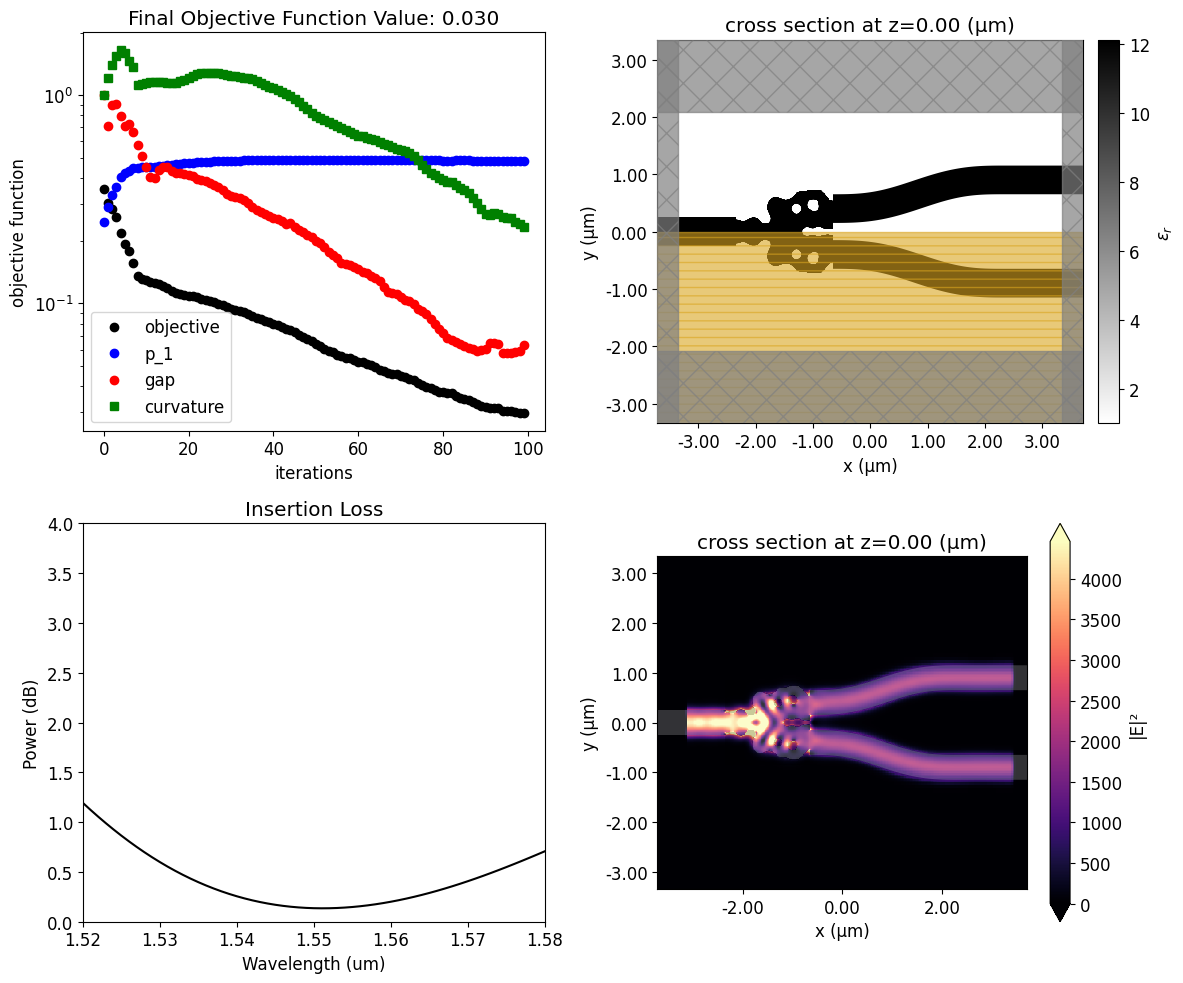
Below, we can see how the device response and fabrication penalty have evolved throughout the optimization. The coupling into the output waveguide improves quickly in the beginning at the expense of higher penalty values. Then, the penalty values decrease linearly after the device response achieves a near-optimal condition. This trend results from the small weight factor we have chosen for the fabrication penalty terms.
[25]:
fig, ax = plt.subplots(1, 1, figsize=(6, 4))
ax.plot(obj_vals, "ko", label="objective")
ax.plot(eta1_vals, "bo", label="p_1")
ax.plot(pen_gap_vals, "ro", label="gap")
ax.plot(pen_curve_vals, "gs", label="curvature")
ax.set_xlabel("iterations")
ax.set_ylabel("objective function")
ax.legend()
ax.set_yscale("log")
ax.set_title(f"Final Objective Function Value: {obj_vals[-1]:.3f}")
plt.show()
The optimization process obtained a smooth and well-defined geometry.
[26]:
eps_final = get_eps(mirror_param(final_par), plot_levelset=True)
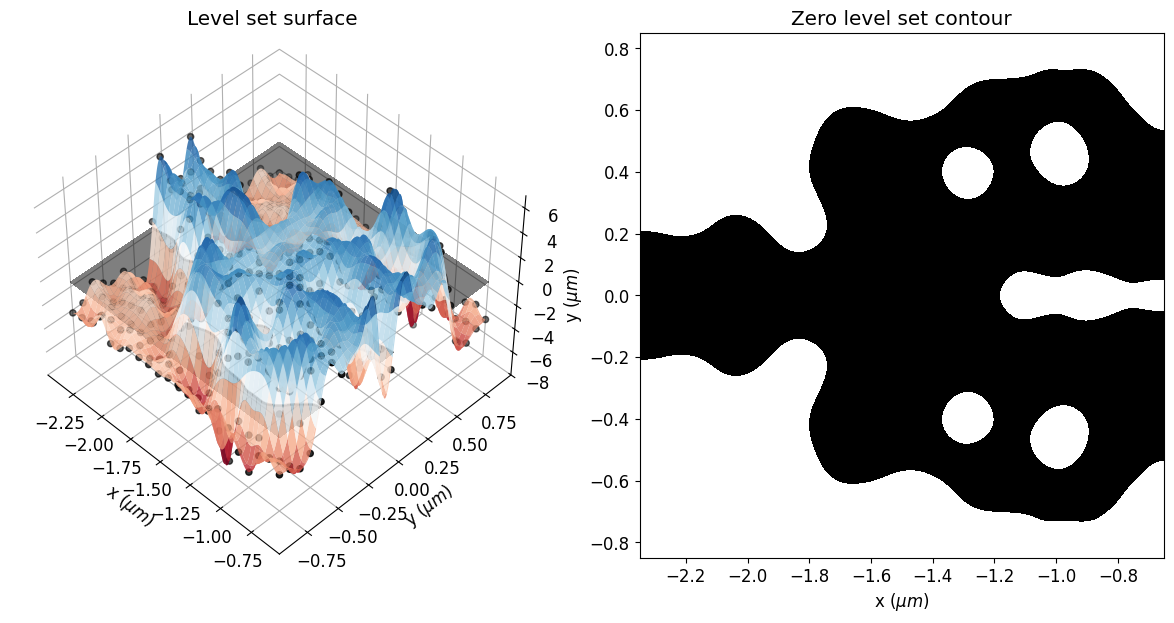
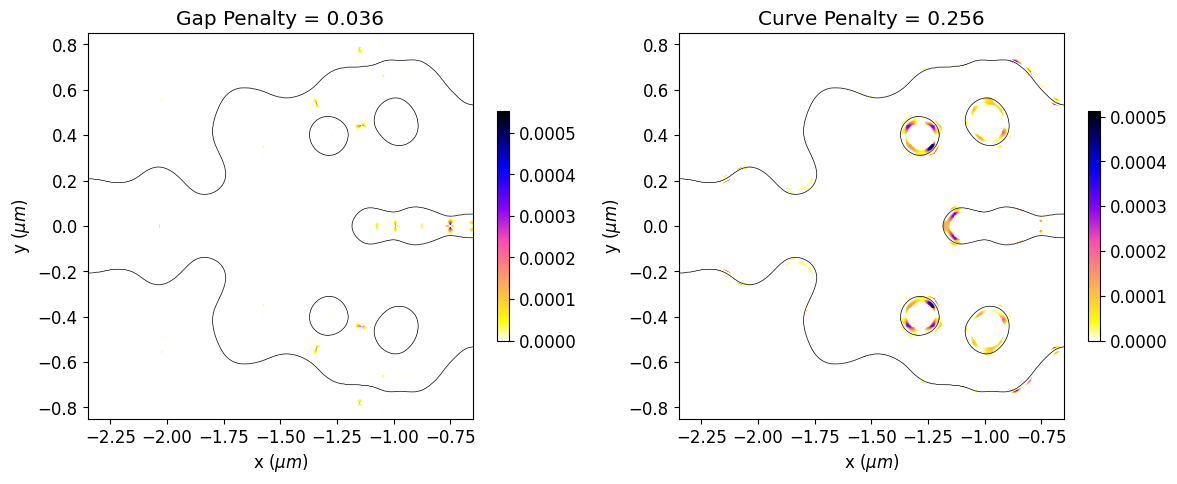
We can also see a significant reduction in violations to the minimum feature size after the optimization, which results in a smoother structure. The optimized device has not matched the minimum feature size exactly. The minimum radius of curvature and gap size are about 30% higher and 20% lower than the reference feature size, respectively. This deviation is expected, as reported in the previous paper. In this regard, running the simulation longer, adjusting the penalty weight or compensating for the reference feature size could improve the results.
[27]:
# Initial values of gap and curvature fabrication constraints.
final_fab_gap, final_fab_curve = fab_penalty_ls(mirror_param(final_par))
# Visualization of gap and curvature fabrication constraints values.
gap_penalty_int = fab_penalty_ls_gap(mirror_param(final_par), beta=gap_par)
curve_penalty_int = fab_penalty_ls_curve(mirror_param(final_par), alpha=curve_par, sharpness=4)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 8), tight_layout=True)
yy, xx = np.meshgrid(y_phi, x_phi)
im = ax1.imshow(
np.flipud(gap_penalty_int.T),
extent=[x_phi[0], x_phi[-1], y_phi[0], y_phi[-1]],
interpolation="none",
cmap="gnuplot2_r",
)
ax1.contour(xx, yy, eps_final, [(eps_min + eps_max) / 2], colors="k", linewidths=0.5)
ax1.set_title(f"Gap Penalty = {final_fab_gap:.3f}")
ax1.set_xlabel(r"x ($\mu m$)")
ax1.set_ylabel(r"y ($\mu m$)")
fig.colorbar(im, ax=ax1, shrink=0.3)
im = ax2.imshow(
anp.flipud(curve_penalty_int.T),
extent=[x_phi[0], x_phi[-1], y_phi[0], y_phi[-1]],
interpolation="none",
cmap="gnuplot2_r",
)
ax2.contour(xx, yy, eps_final, [(eps_min + eps_max) / 2], colors="k", linewidths=0.5)
ax2.set_title(f"Curve Penalty = {final_fab_curve:.3f}")
ax2.set_xlabel(r"x ($\mu m$)")
ax2.set_ylabel(r"y ($\mu m$)")
fig.colorbar(im, ax=ax2, shrink=0.3)
plt.show()
Once the inverse design is complete, we can visualize the field distributions and the wavelength dependent insertion loss.
[28]:
sim_final = make_adjoint_sim(mirror_param(final_par), unfold=True)
sim_final = sim_final.copy(update=dict(monitors=(field_xy, fom_final_1)))
sim_data_final = web.run(sim_final, task_name="inv_des_final")
08:48:43 CEST Created task 'inv_des_final' with task_id 'fdve-cbf9a3ac-fa64-4092-abea-8024c43fef96' and task_type 'FDTD'.
View task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-cbf9a3ac-fa 64-4092-abea-8024c43fef96'.
Task folder: 'default'.
08:48:46 CEST Maximum FlexCredit cost: 0.025. Minimum cost depends on task execution details. Use 'web.real_cost(task_id)' to get the billed FlexCredit cost after a simulation run.
08:48:47 CEST status = queued
To cancel the simulation, use 'web.abort(task_id)' or 'web.delete(task_id)' or abort/delete the task in the web UI. Terminating the Python script will not stop the job running on the cloud.
08:49:01 CEST status = preprocess
08:49:05 CEST starting up solver
running solver
08:49:11 CEST early shutoff detected at 60%, exiting.
08:49:12 CEST status = postprocess
08:49:14 CEST status = success
08:49:16 CEST View simulation result at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-cbf9a3ac-fa 64-4092-abea-8024c43fef96'.
08:49:18 CEST loading simulation from simulation_data.hdf5
The resulting structure shows good performance, presenting insertion loss of only 0.1 dB near the central wavelength.
[29]:
mode_amps = sim_data_final["out_1"]
coeffs_f = mode_amps.amps.sel(direction="+")
power_1 = np.abs(coeffs_f.sel(mode_index=0)) ** 2
power_1_db = -10 * np.log10(2 * power_1)
f, ax = plt.subplots(2, 2, figsize=(12, 10), tight_layout=True)
sim_final.plot_eps(z=0, source_alpha=0, monitor_alpha=0, ax=ax[0, 1])
ax[1, 0].plot(wl_range, power_1_db, "-k")
ax[1, 0].set_xlabel("Wavelength (um)")
ax[1, 0].set_ylabel("Power (dB)")
ax[1, 0].set_ylim(0, 4)
ax[1, 0].set_xlim(wl - bw / 2, wl + bw / 2)
ax[1, 0].set_title("Insertion Loss")
sim_data_final.plot_field("field_xy", "E", "abs^2", z=0, ax=ax[1, 1])
ax[0, 0].plot(obj_vals, "ko", label="objective")
ax[0, 0].plot(eta1_vals, "bo", label="p_1")
ax[0, 0].plot(pen_gap_vals, "ro", label="gap")
ax[0, 0].plot(pen_curve_vals, "gs", label="curvature")
ax[0, 0].set_xlabel("iterations")
ax[0, 0].set_ylabel("objective function")
ax[0, 0].legend()
ax[0, 0].set_yscale("log")
ax[0, 0].set_title(f"Final Objective Function Value: {obj_vals[-1]:.3f}")
plt.show()
Export to GDS#
The Simulation object has the .to_gds_file convenience function to export the final design to a GDS file. In addition to a file name, it is necessary to set a cross-sectional plane (z = 0 in this case) on which to evaluate the geometry, a frequency to evaluate the permittivity, and a permittivity_threshold to define the shape boundaries in custom
mediums. See the GDS export notebook for a detailed example on using .to_gds_file and other GDS related functions.
[30]:
sim_final.to_gds_file(
fname="./misc/inv_des_ybranch.gds",
z=0,
permittivity_threshold=(eps_max + eps_min) / 2,
frequency=freq,
)