Adjoint inverse design of a quantum emitter light extractor#
The cost of running the entire optimization is about 8 FlexCredit (check this)
In this tutorial, we will show how to perform the adjoint-based inverse design of a quantum emitter (QE) light extraction structure. We will use a PointDipole to model the QE embedded within an integrated dielectric waveguide. Then, we will build an optimization problem to maximize the extraction efficiency of the dipole radiation into a collection waveguide. In addition, we will show how to use FieldMonitor objects in adjoint simulations to calculate the flux radiated from the dipole.
You can also find helpful information in this related notebook.
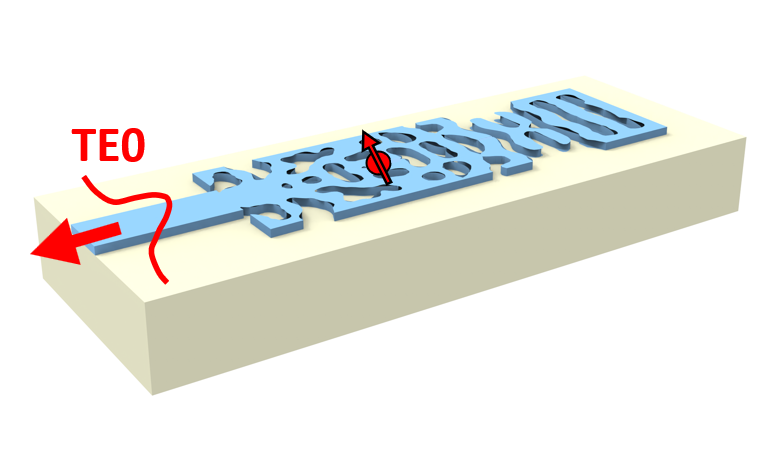
If you are unfamiliar with inverse design, we recommend the inverse design lectures and this introductory tutorial.
Let’s start by importing the Python libraries used throughout this notebook.
[1]:
# Standard python imports.
import pickle
from typing import List
# Import autograd for automatic differentiation.
import autograd as ag
import autograd.numpy as anp
import matplotlib.pylab as plt
import numpy as np
import optax
import scipy as sp
# Import regular tidy3d.
import tidy3d as td
import tidy3d.web as web
from tidy3d.plugins.autograd import (
make_erosion_dilation_penalty,
make_filter_and_project,
rescale,
value_and_grad,
)
Simulation Set Up#
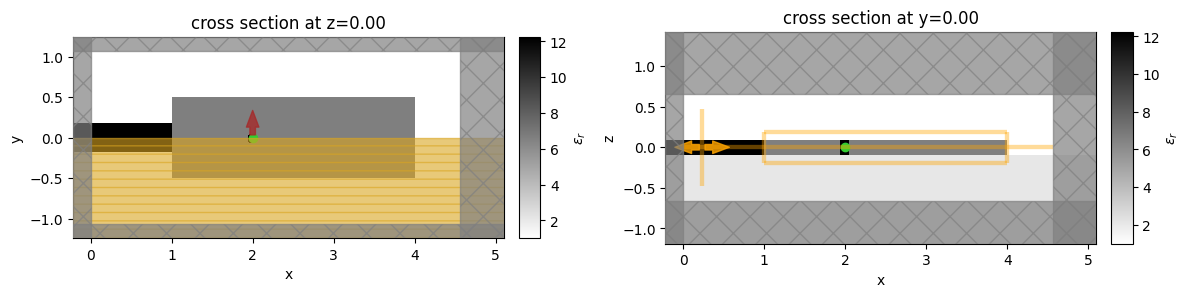
The coupling region (design region) extends a single-mode dielectric waveguide placed over a lower refractive index substrate. The QE is modeled as a PointDipole oriented in the y-direction. The QE is placed within the design region so we surround it with a constant refractive index region to protect it from etching.
[2]:
# Geometric parameters.
cr_w = 1.0 # Coupling region width (um).
cr_l = 3.0 # Coupling region length (um).
wg_thick = 0.19 # Collection waveguide thickness (um).
wg_width = 0.35 # Collection waveguide width (um).
wg_length = 1.0 # Collection waveguide length (um).
# Material.
n_wg = 3.50 # Structure refractive index.
n_sub = 1.44 # Substrate refractive index.
# Fabrication constraints.
min_feature = 0.06 # Minimum feature size.
non_etch_r = 0.06 # Non-etched circular region radius (um).
# Inverse design set up parameters.
grid_size = 0.015 # Simulation grid size on design region (um).
max_iter = 100 # Maximum number of iterations.
iter_steps = 5 # Beta is increased at each iter_steps.
beta_min = 1.0 # Minimum value for the tanh projection parameter.
learning_rate = 0.02
# Simulation wavelength.
wl = 0.94 # Central simulation wavelength (um).
bw = 0.04 # Simulation bandwidth (um).
n_wl = 41 # Number of wavelength points within the bandwidth.
Let’s calculate some variables used throughout the notebook. Here, we will also define the QE position and monitor planes.
[3]:
# Minimum and maximum values of the permittivity.
eps_max = n_wg**2
eps_min = 1.0
# Material definition.
mat_wg = td.Medium(permittivity=eps_max)
mat_sub = td.Medium(permittivity=n_sub**2)
# Wavelengths and frequencies.
wl_max = wl + bw / 2
wl_min = wl - bw / 2
wl_range = np.linspace(wl_min, wl_max, n_wl)
freq = td.C_0 / wl
freqs = td.C_0 / wl_range
freqw = 0.5 * (freqs[0] - freqs[-1])
run_time = 3e-12
# Computational domain size.
pml_spacing = 0.6 * wl
size_x = wg_length + cr_l + pml_spacing
size_y = cr_w + 2 * pml_spacing
size_z = wg_thick + 2 * pml_spacing
eff_inf = 10
# Source position and monitor planes.
cr_center_x = wg_length + cr_l / 2
qe_pos = td.Box(center=(cr_center_x - 0.5, 0, 0), size=(0, 0, 0))
qe_field_plan = td.Box.surfaces(center=(cr_center_x, 0, 0), size=(cr_l, cr_w, 2 * wg_thick))
wg_mode_plan = td.Box(center=(wl / 4, 0, 0), size=(0, 4 * wg_width, 5 * wg_thick))
# Number of points on design grid.
nx_grid = int(cr_l / grid_size)
ny_grid = int(cr_w / grid_size / 2)
# xy coordinates of design grid.
x_grid = np.linspace(cr_center_x - cr_l / 2, cr_center_x + cr_l / 2, nx_grid)
y_grid = np.linspace(0, cr_w / 2, ny_grid)
Optimization Set Up#
We will start defining the density-based optimization functions to transform the design parameters into permittivity values. Here we include the ConicFilter, where we impose a minimum feature size fabrication constraint, and the tangent hyperbolic projection function, eliminating intermediary permittivity values as we increase the projection parameter beta. You can find more information in the Inverse design optimization of a compact grating
coupler.
[4]:
def pre_process(params, beta):
filter_project = make_filter_and_project(
radius=min_feature, dl=grid_size, beta=beta, eta=0.5, filter_type="conic"
)
params1 = filter_project(params, beta)
return params1
def get_eps(params, beta: float = 1.00):
"""Returns the permittivities after filter and projection transformations"""
params1 = pre_process(params, beta=beta)
eps = eps_min + (eps_max - eps_min) * params1
eps = anp.maximum(eps, eps_min)
eps = anp.minimum(eps, eps_max)
return eps
This function includes a circular region of constant permittivity value surrounding the QE. The objective here is to protect the QE from etching. In applications such as single photon sources, a larger unperturbed region surrounding the QE can be helpful to reduce linewidth broadening, as stated in
J. Liu, K. Konthasinghe, M. Davanco, J. Lawall, V. Anant, V. Verma, R. Mirin, S. Nam, S. Woo, D. Jin, B. Ma, Z. Chen, H. Ni, Z. Niu, K. Srinivasan, "Single Self-Assembled InAs/GaAs Quantum Dots in Photonic Nanostructures: The Role of Nanofabrication," Phys. Rev. Appl. 9(6), 064019 (2018) DOI: 10.1103/PhysRevApplied.9.064019.
[5]:
def include_constant_regions(eps, circ_center=[0, 0], circ_radius=1.0):
# Build the geometric mask.
yv, xv = anp.meshgrid(y_grid, x_grid)
# Shouldn't this be --> |x-x0|^2 + |y-y0|^2 <= r*2
geo_mask = (
anp.where(
anp.abs((xv - circ_center[0]) ** 2 + (yv - circ_center[1]) ** 2) <= (circ_radius**2),
1,
0,
)
* eps_max
)
eps = anp.maximum(geo_mask, eps)
return eps
Now, we define a function to update the td.CustomMedium using the permittivity distribution. The simulation will include mirror symmetry concerning the y-direction, so only the upper half of the design region is returned by this function during the optimization process. To get the whole structure, you need to set unfold=True.
[6]:
def update_design(eps, unfold=False) -> List[td.Structure]:
# Definition of the coordinates x,y along the design region.
coords_x = [(cr_center_x - cr_l / 2) + ix * grid_size for ix in range(nx_grid)]
eps_val = anp.array(eps).reshape((nx_grid, ny_grid, 1))
if not unfold:
coords_yp = [0 + iy * grid_size for iy in range(ny_grid)]
coords = dict(x=coords_x, y=coords_yp, z=[0])
eps1 = td.SpatialDataArray(eps_val, coords)
eps_medium = td.CustomMedium(permittivity=eps1)
box = td.Box(center=(cr_center_x, cr_w / 4, 0), size=(cr_l, cr_w / 2, wg_thick))
structure = [td.Structure(geometry=box, medium=eps_medium)]
# VJP for one of anp.copy(), anp.concatenate(), or anp.fliplr() not defined,
# so the optimization should only be run with `unfold=False` for now
else:
coords_y = [-cr_w / 2 + iy * grid_size for iy in range(2 * ny_grid)]
coords = dict(x=coords_x, y=coords_y, z=[0])
eps1 = td.SpatialDataArray(
anp.concatenate((anp.fliplr(anp.copy(eps_val)), eps_val), axis=1), coords
)
eps_medium = td.CustomMedium(permittivity=eps1)
box = td.Box(center=(cr_center_x, 0, 0), size=(cr_l, cr_w, wg_thick))
structure = [td.Structure(geometry=box, medium=eps_medium)]
return structure
In the next cell, we define the output waveguide and the substrate, as well as the simulation monitors. It is worth mentioning the inclusion of a ModeMonitor in the output waveguide and a FieldMonitor box surrounding the dipole source to calculate the total radiated power.
[7]:
# Input/output waveguide.
waveguide = td.Structure(
geometry=td.Box.from_bounds(
rmin=(-eff_inf, -wg_width / 2, -wg_thick / 2),
rmax=(wg_length, wg_width / 2, wg_thick / 2),
),
medium=mat_wg,
)
# Substrate layer.
substrate = td.Structure(
geometry=td.Box.from_bounds(
rmin=(-eff_inf, -eff_inf, -eff_inf), rmax=(eff_inf, eff_inf, -wg_thick / 2)
),
medium=mat_sub,
)
# Point dipole source located at the center of TiO2 thin film.
dp_source = td.PointDipole(
center=qe_pos.center,
source_time=td.GaussianPulse(freq0=freq, fwidth=freqw),
polarization="Ey",
)
# Mode monitor to compute the FOM.
mode_spec = td.ModeSpec(num_modes=1, target_neff=n_wg)
mode_monitor_fom = td.ModeMonitor(
center=wg_mode_plan.center,
size=wg_mode_plan.size,
freqs=[freq],
mode_spec=mode_spec,
name="mode_monitor_fom",
)
# Field monitor to compute the FOM.
field_monitor_fom = []
for i, plane in enumerate(qe_field_plan):
field_monitor_fom.append(
td.FieldMonitor(
center=plane.center,
size=plane.size,
freqs=[freq],
name=f"field_monitor_fom_{i}",
)
)
# Mode monitor to compute spectral response.
mode_spec = td.ModeSpec(num_modes=1, target_neff=n_wg)
mode_monitor = td.ModeMonitor(
center=wg_mode_plan.center,
size=wg_mode_plan.size,
freqs=freqs,
mode_spec=mode_spec,
name="mode_monitor",
)
# Field monitor to compute spectral response.
field_monitor = []
for i, plane in enumerate(qe_field_plan):
field_monitor.append(
td.FieldMonitor(
center=plane.center, size=plane.size, freqs=freqs, name=f"field_monitor_{i}"
)
)
# Field monitor to visualize the fields.
field_monitor_xy = td.FieldMonitor(
center=(size_x / 2, 0, 0),
size=(size_x, size_y, 0),
freqs=freqs,
name="field_xy",
)
Lastly, we have a function that receives the design parameters from the optimization algorithm and then gathers the simulation objects altogether to create a td.Simulation.
[8]:
def make_adjoint_sim(param, beta: float = 1.00, unfold=False):
eps = get_eps(param, beta)
eps = include_constant_regions(
eps, circ_center=[qe_pos.center[0], qe_pos.center[1]], circ_radius=non_etch_r
)
structure = update_design(eps, unfold=unfold)
# Creates a uniform mesh for the design region.
adjoint_dr_mesh = td.MeshOverrideStructure(
geometry=td.Box(center=(cr_center_x, 0, 0), size=(cr_w, cr_l, wg_thick)),
dl=[grid_size, grid_size, grid_size],
enforce=True,
)
grid_spec = td.GridSpec.auto(
wavelength=wl_max,
min_steps_per_wvl=15,
override_structures=[adjoint_dr_mesh],
)
return td.Simulation(
size=[size_x, size_y, size_z],
center=[size_x / 2, 0, 0],
grid_spec=grid_spec,
symmetry=(0, -1, 0),
structures=[substrate, waveguide] + structure,
sources=[dp_source],
monitors=[field_monitor_xy, mode_monitor_fom] + field_monitor_fom,
run_time=run_time,
subpixel=True,
)
Initial Light Extractor Structure#
Let’s create a uniform initial permittivity distribution and verify if all the simulation objects are in the correct places.
[9]:
init_par = np.ones((nx_grid, ny_grid)) * 0.5
init_design = make_adjoint_sim(init_par, beta=beta_min, unfold=True)
fig, (ax1, ax2) = plt.subplots(1, 2, tight_layout=True, figsize=(12, 4))
init_design.plot_eps(z=0, ax=ax1, monitor_alpha=0.0)
init_design.plot_eps(y=0, ax=ax2)
plt.show()
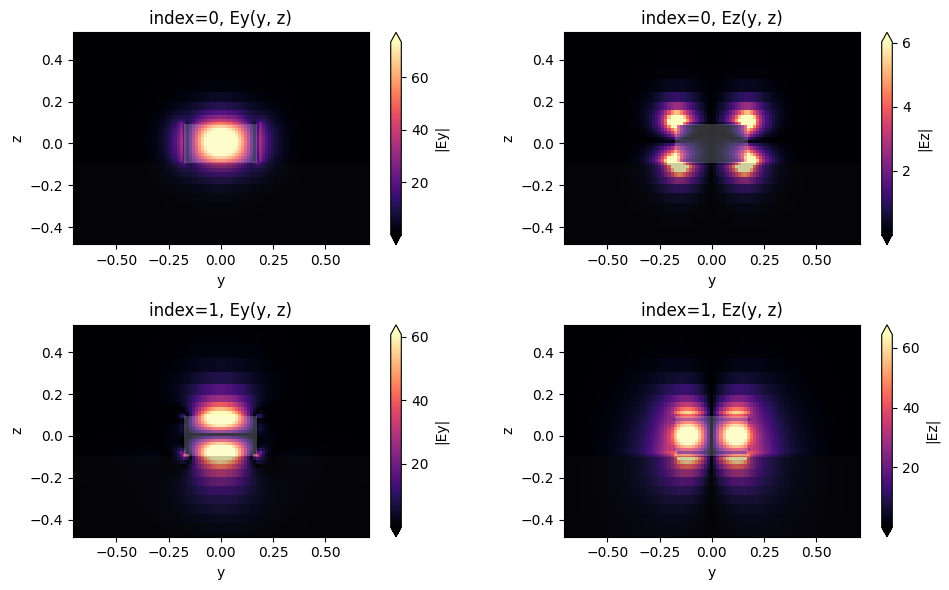
We will also look at the collection waveguide mode to ensure we have considered the correct one in the ModeMonitor setup. We use the ModeSolver plugin to calculate the first two waveguide modes, as below.
[10]:
from tidy3d.plugins.mode import ModeSolver
from tidy3d.plugins.mode.web import run as run_mode_solver
sim_init = init_design.updated_copy(monitors=[field_monitor_xy, mode_monitor] + field_monitor)
mode_solver = ModeSolver(
simulation=sim_init,
plane=wg_mode_plan,
mode_spec=td.ModeSpec(num_modes=2),
freqs=[freq],
)
modes = run_mode_solver(mode_solver, reduce_simulation=True)
16:10:23 CEST Mode solver created with task_id='fdve-65e65354-aedf-458c-a514-a7eccc5e4b24', solver_id='mo-581ee868-4940-489c-bab0-2c43f11c8df2'.
16:10:26 CEST Mode solver status: queued
16:10:28 CEST Mode solver status: running
16:10:32 CEST Mode solver status: success
After inspecting the mode field distribution, we can confirm that the fundamental waveguide mode is mainly oriented in the y-direction, thus matching the dipole orientation.
[11]:
fig, axs = plt.subplots(2, 2, figsize=(10, 6), tight_layout=True)
for mode_ind in range(2):
for field_ind, field_name in enumerate(("Ey", "Ez")):
ax = axs[mode_ind, field_ind]
mode_solver.plot_field(field_name, "abs", mode_index=mode_ind, f=freq, ax=ax)
ax.set_title(f"index={mode_ind}, {field_name}(y, z)")
Then, we will calculate the initial coupling efficiency to see how this random structure performs.
[12]:
sim_data = web.run(sim_init, task_name="initial QE light extractor (Autograd)")
16:10:35 CEST Created task 'initial QE light extractor (Autograd)' with task_id 'fdve-4ca772ae-badc-4827-8da8-4b32cf325ab4' and task_type 'FDTD'.
View task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-4ca772ae-ba dc-4827-8da8-4b32cf325ab4'.
Task folder: 'default'.
16:10:37 CEST Maximum FlexCredit cost: 0.060. Minimum cost depends on task execution details. Use 'web.real_cost(task_id)' to get the billed FlexCredit cost after a simulation run.
16:10:38 CEST status = success
16:10:43 CEST loading simulation from simulation_data.hdf5
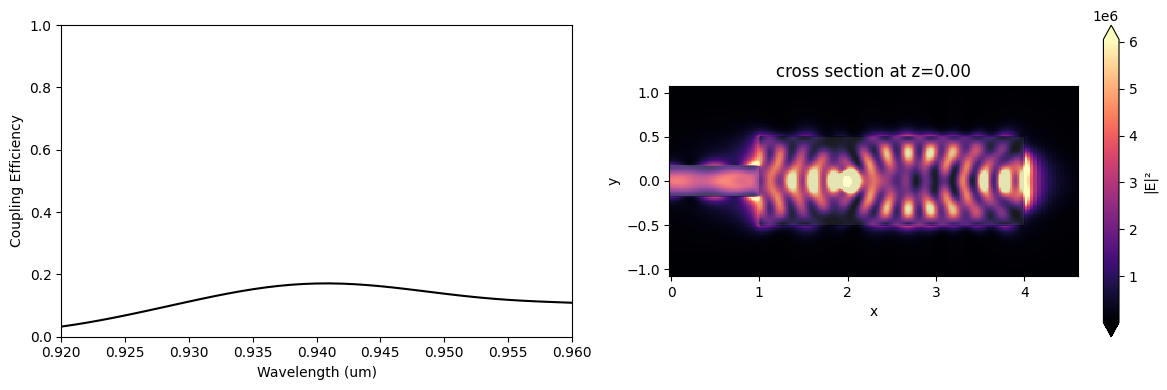
The modal coupling efficiency is normalized by the dipole power. That is necessary because the dipole power will likely change significantly when the optimization algorithm modifies the design region.
[13]:
mode_amps = sim_data["mode_monitor"].amps.sel(direction="-", mode_index=0)
mode_power = np.abs(mode_amps) ** 2
dip_power = np.zeros(n_wl)
for i in range(len(field_monitor)):
field_mon = sim_data[f"field_monitor_{i}"]
dip_power += np.abs(field_mon.flux)
coup_eff = mode_power / dip_power
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4), tight_layout=True)
ax1.plot(wl_range, coup_eff, "-k")
ax1.set_xlabel("Wavelength (um)")
ax1.set_ylabel("Coupling Efficiency")
ax1.set_ylim(0, 1)
ax1.set_xlim(wl - bw / 2, wl + bw / 2)
sim_data.plot_field("field_xy", "E", "abs^2", z=0, ax=ax2, f=freq)
plt.show()
Optimization#
The objective function defined next is the device figure-of-merit (FOM) minus a fabrication penalty.
[14]:
# Figure of Merit (FOM) calculation.
def fom(sim_data: td.SimulationData) -> float:
"""Return the coupling efficiency."""
# best to use autograd-wrapped numpy functions for differentiation
mode_amps = sim_data["mode_monitor_fom"].amps.sel(direction="-", f=freq, mode_index=0).data
mode_power = anp.sum(anp.abs(mode_amps) ** 2)
# unlike Jax version, should avoid in-place operators (e.g, `+=`), use numpy when possible
field_mon_list = [sim_data[f"field_monitor_fom_{i}"] for i in range(0, 6)]
dip_power = anp.sum([anp.abs(mon.flux.data) for mon in field_mon_list])
return mode_power, dip_power
def penalty(params, beta) -> float:
"""Penalize changes in structure after erosion and dilation to enforce larger feature sizes."""
params_processed = pre_process(params, beta=beta)
erode_dilate_penalty = make_erosion_dilation_penalty(radius=min_feature, dl=grid_size)
ed_penalty = erode_dilate_penalty(params_processed)
return ed_penalty
# Objective function to be passed to the optimization algorithm.
def obj(param, beta: float = 1.0, step_num: int = None, verbose: bool = False) -> float:
sim = make_adjoint_sim(param, beta, unfold=False) # non-differentiable if `unfold=True`
task_name = "inv_des"
if step_num:
task_name += f"_step_{step_num}"
sim_data = web.run(sim, task_name=task_name, verbose=verbose)
mode_power, dip_power = fom(sim_data)
fom_val = mode_power / dip_power
penalty_weight = 0.1
penalty_val = penalty(param, beta)
J = fom_val - penalty_weight * penalty_val
return J, [sim_data, mode_power, dip_power, penalty_val]
# Function to calculate the objective function value and its gradient with respect to the design parameters.
# Use tidy3d's wrapped ag.value_and_grad() for it's auxiliary data functionality
obj_grad = value_and_grad(obj, has_aux=True)
In the following cell, we define some functions to save the optimization progress and load a previous optimization from the file.
[15]:
# where to store history
history_fname = "misc/qe_light_coupler_autograd.pkl"
def save_history(history_dict: dict) -> None:
"""Convenience function to save the history to file."""
with open(history_fname, "wb") as file:
pickle.dump(history_dict, file)
def load_history() -> dict:
"""Convenience method to load the history from file."""
with open(history_fname, "rb") as file:
history_dict = pickle.load(file)
return history_dict
Then, we will start a new optimization or load the parameters of a previous one.
[16]:
# initialize adam optimizer with starting parameters
optimizer = optax.adam(learning_rate=learning_rate)
try:
history_dict = load_history()
opt_state = history_dict["opt_states"][-1]
params = history_dict["params"][-1]
opt_state = optimizer.init(params)
num_iters_completed = len(history_dict["params"])
print("Loaded optimization checkpoint from file.")
print(f"Found {num_iters_completed} iterations previously completed out of {max_iter} total.")
if num_iters_completed < max_iter:
print("Will resume optimization.")
else:
print("Optimization completed, will return results.")
except FileNotFoundError:
params = anp.array(init_par)
opt_state = optimizer.init(params)
history_dict = dict(
values=[],
coupl_eff=[],
penalty=[],
params=[],
gradients=[],
opt_states=[opt_state],
data=[],
beta=[],
)
WARNING:2025-07-26 16:10:43,499:jax._src.xla_bridge:967: An NVIDIA GPU may be present on this machine, but a CUDA-enabled jaxlib is not installed. Falling back to cpu.
In the optimization loop, we will gradually increase the projection parameter beta to eliminate intermediary permittivity values. At each iteration, we record the design parameters and the optimization history to restore them as needed.
[17]:
iter_done = len(history_dict["values"])
if iter_done < max_iter:
# small # of iters for quick testing
for i in range(iter_done, max_iter):
print(f"Iteration = ({i + 1} / {max_iter})")
plt.subplots(1, 1, figsize=(3, 2))
plt.imshow(np.flipud(1 - params.T), cmap="gray", vmin=0, vmax=1)
plt.axis("off")
plt.show()
# Compute gradient and current objective function value.
beta_i = i // iter_steps + beta_min
(value, gradient), data = obj_grad(params, beta=beta_i, step_num=(i + 1))
sim_data_i, mode_power_i, dip_power_i, penalty_val_i = [data[0]] + [
dat._value for dat in data[1:]
]
# Outputs.
print(f"\tbeta = {beta_i}")
print(f"\tJ = {value:.4e}")
print(f"\tgrad_norm = {np.linalg.norm(gradient):.4e}")
print(f"\tpenalty = {penalty_val_i:.3f}")
print(f"\tmode power = {mode_power_i:.3f}")
print(f"\tdip power = {dip_power_i:.3f}")
print(f"\tcoupling efficiency = {mode_power_i / dip_power_i:.3f}")
# Compute and apply updates to the optimizer based on gradient (-1 sign to maximize obj_fn).
updates, opt_state = optimizer.update(-gradient, opt_state, params)
params = optax.apply_updates(params, updates)
# Cap parameters between 0 and 1.
params = anp.minimum(params, 1.0)
params = anp.maximum(params, 0.0)
# Save history.
history_dict["values"].append(value)
history_dict["coupl_eff"].append(mode_power_i / dip_power_i)
history_dict["penalty"].append(penalty_val_i)
history_dict["params"].append(params)
history_dict["beta"].append(beta_i)
history_dict["gradients"].append(gradient)
history_dict["opt_states"].append(opt_state)
# history_dict["data"].append(sim_data_i) # Uncomment to store data, can create large files.
save_history(history_dict)
Iteration = (1 / 100)

beta = 1.0
J = 7.1432e-02
grad_norm = 6.8739e-02
penalty = 1.000
mode power = 473.968
dip power = 2764.752
coupling efficiency = 0.171
Iteration = (2 / 100)

beta = 1.0
J = 2.2832e-01
grad_norm = 7.1512e-02
penalty = 1.000
mode power = 1002.394
dip power = 3053.089
coupling efficiency = 0.328
Iteration = (3 / 100)

beta = 1.0
J = 3.7482e-01
grad_norm = 6.4898e-02
penalty = 1.000
mode power = 1478.833
dip power = 3114.501
coupling efficiency = 0.475
Iteration = (4 / 100)

beta = 1.0
J = 4.9571e-01
grad_norm = 5.6425e-02
penalty = 1.000
mode power = 1935.575
dip power = 3249.175
coupling efficiency = 0.596
Iteration = (5 / 100)

beta = 1.0
J = 5.8980e-01
grad_norm = 4.6824e-02
penalty = 1.000
mode power = 2418.556
dip power = 3506.185
coupling efficiency = 0.690
Iteration = (6 / 100)

beta = 2.0
J = 6.9596e-01
grad_norm = 5.4160e-02
penalty = 1.000
mode power = 3059.636
dip power = 3843.981
coupling efficiency = 0.796
Iteration = (7 / 100)

beta = 2.0
J = 7.4070e-01
grad_norm = 3.8978e-02
penalty = 1.000
mode power = 3716.945
dip power = 4421.261
coupling efficiency = 0.841
Iteration = (8 / 100)

beta = 2.0
J = 7.7138e-01
grad_norm = 3.0304e-02
penalty = 1.000
mode power = 4429.873
dip power = 5083.755
coupling efficiency = 0.871
Iteration = (9 / 100)

beta = 2.0
J = 7.9249e-01
grad_norm = 2.9056e-02
penalty = 1.000
mode power = 5169.262
dip power = 5791.995
coupling efficiency = 0.892
Iteration = (10 / 100)

beta = 2.0
J = 8.0856e-01
grad_norm = 2.8136e-02
penalty = 1.000
mode power = 6042.956
dip power = 6651.128
coupling efficiency = 0.909
Iteration = (11 / 100)

beta = 3.0
J = 7.9742e-01
grad_norm = 9.7527e-02
penalty = 1.000
mode power = 7003.191
dip power = 7803.999
coupling efficiency = 0.897
Iteration = (12 / 100)

beta = 3.0
J = 8.3259e-01
grad_norm = 3.7221e-02
penalty = 0.999
mode power = 8407.922
dip power = 9016.226
coupling efficiency = 0.933
Iteration = (13 / 100)
beta = 3.0
J = 8.2696e-01
grad_norm = 6.7464e-02
penalty = 0.999
mode power = 8736.356
dip power = 9425.743
coupling efficiency = 0.927
Iteration = (14 / 100)
beta = 3.0
J = 8.1778e-01
grad_norm = 1.0335e-01
penalty = 0.999
mode power = 8016.955
dip power = 8736.591
coupling efficiency = 0.918
Iteration = (15 / 100)
beta = 3.0
J = 8.3701e-01
grad_norm = 7.7907e-02
penalty = 0.998
mode power = 7685.024
dip power = 8203.539
coupling efficiency = 0.937
Iteration = (16 / 100)
beta = 4.0
J = 8.1204e-01
grad_norm = 1.5668e-01
penalty = 0.983
mode power = 5820.384
dip power = 6393.784
coupling efficiency = 0.910
Iteration = (17 / 100)
beta = 4.0
J = 8.4011e-01
grad_norm = 1.0337e-01
penalty = 0.977
mode power = 7501.458
dip power = 7998.690
coupling efficiency = 0.938
Iteration = (18 / 100)
beta = 4.0
J = 8.6124e-01
grad_norm = 2.8364e-02
penalty = 0.971
mode power = 10468.018
dip power = 10923.463
coupling efficiency = 0.958
Iteration = (19 / 100)
beta = 4.0
J = 8.5361e-01
grad_norm = 7.5763e-02
penalty = 0.963
mode power = 13775.781
dip power = 14501.615
coupling efficiency = 0.950
Iteration = (20 / 100)
beta = 4.0
J = 8.5295e-01
grad_norm = 8.1482e-02
penalty = 0.956
mode power = 16346.014
dip power = 17233.051
coupling efficiency = 0.949
Iteration = (21 / 100)
beta = 5.0
J = 8.7218e-01
grad_norm = 5.7052e-02
penalty = 0.875
mode power = 17111.887
dip power = 17829.989
coupling efficiency = 0.960
Iteration = (22 / 100)
beta = 5.0
J = 8.8005e-01
grad_norm = 5.3833e-02
penalty = 0.865
mode power = 17308.501
dip power = 17908.051
coupling efficiency = 0.967
Iteration = (23 / 100)
beta = 5.0
J = 8.8289e-01
grad_norm = 3.2505e-02
penalty = 0.854
mode power = 15082.259
dip power = 15576.252
coupling efficiency = 0.968
Iteration = (24 / 100)
beta = 5.0
J = 8.8113e-01
grad_norm = 4.4331e-02
penalty = 0.843
mode power = 12419.322
dip power = 12863.968
coupling efficiency = 0.965
Iteration = (25 / 100)
beta = 5.0
J = 8.7947e-01
grad_norm = 6.3651e-02
penalty = 0.832
mode power = 12087.040
dip power = 12555.809
coupling efficiency = 0.963
Iteration = (26 / 100)
beta = 6.0
J = 8.8584e-01
grad_norm = 7.8859e-02
penalty = 0.732
mode power = 10924.212
dip power = 11390.265
coupling efficiency = 0.959
Iteration = (27 / 100)
beta = 6.0
J = 8.9808e-01
grad_norm = 3.3468e-02
penalty = 0.722
mode power = 17336.079
dip power = 17867.154
coupling efficiency = 0.970
Iteration = (28 / 100)
beta = 6.0
J = 9.0230e-01
grad_norm = 7.8044e-02
penalty = 0.712
mode power = 25590.844
dip power = 26288.327
coupling efficiency = 0.973
Iteration = (29 / 100)
beta = 6.0
J = 8.9802e-01
grad_norm = 9.3922e-02
penalty = 0.702
mode power = 26559.148
dip power = 27429.898
coupling efficiency = 0.968
Iteration = (30 / 100)
beta = 6.0
J = 8.9534e-01
grad_norm = 7.9459e-02
penalty = 0.694
mode power = 21200.802
dip power = 21975.043
coupling efficiency = 0.965
Iteration = (31 / 100)
beta = 7.0
J = 8.9894e-01
grad_norm = 1.4495e-01
penalty = 0.610
mode power = 11024.127
dip power = 11483.994
coupling efficiency = 0.960
Iteration = (32 / 100)
beta = 7.0
J = 9.0821e-01
grad_norm = 7.0287e-02
penalty = 0.604
mode power = 15566.298
dip power = 16071.417
coupling efficiency = 0.969
Iteration = (33 / 100)
beta = 7.0
J = 9.0284e-01
grad_norm = 1.1942e-01
penalty = 0.597
mode power = 24449.286
dip power = 25399.992
coupling efficiency = 0.963
Iteration = (34 / 100)
beta = 7.0
J = 9.0576e-01
grad_norm = 1.2081e-01
penalty = 0.591
mode power = 25232.320
dip power = 26150.739
coupling efficiency = 0.965
Iteration = (35 / 100)
beta = 7.0
J = 9.1589e-01
grad_norm = 4.2125e-02
penalty = 0.585
mode power = 18147.493
dip power = 18623.626
coupling efficiency = 0.974
Iteration = (36 / 100)
beta = 8.0
J = 9.1102e-01
grad_norm = 1.3288e-01
penalty = 0.522
mode power = 8422.290
dip power = 8743.769
coupling efficiency = 0.963
Iteration = (37 / 100)
beta = 8.0
J = 9.1466e-01
grad_norm = 1.2887e-01
penalty = 0.518
mode power = 11193.142
dip power = 11582.000
coupling efficiency = 0.966
Iteration = (38 / 100)
beta = 8.0
J = 9.2233e-01
grad_norm = 4.5024e-02
penalty = 0.514
mode power = 22909.328
dip power = 23528.628
coupling efficiency = 0.974
Iteration = (39 / 100)
beta = 8.0
J = 9.1957e-01
grad_norm = 1.8263e-01
penalty = 0.510
mode power = 33641.288
dip power = 34663.037
coupling efficiency = 0.971
Iteration = (40 / 100)
beta = 8.0
J = 9.2188e-01
grad_norm = 9.1393e-02
penalty = 0.505
mode power = 29094.673
dip power = 29920.135
coupling efficiency = 0.972
Iteration = (41 / 100)
beta = 9.0
J = 9.1973e-01
grad_norm = 1.7000e-01
penalty = 0.458
mode power = 10998.133
dip power = 11390.796
coupling efficiency = 0.966
Iteration = (42 / 100)
beta = 9.0
J = 9.2859e-01
grad_norm = 9.8428e-02
penalty = 0.454
mode power = 11288.366
dip power = 11589.890
coupling efficiency = 0.974
Iteration = (43 / 100)
beta = 9.0
J = 9.2298e-01
grad_norm = 1.3906e-01
penalty = 0.450
mode power = 16924.833
dip power = 17484.737
coupling efficiency = 0.968
Iteration = (44 / 100)
beta = 9.0
J = 9.2668e-01
grad_norm = 1.0958e-01
penalty = 0.446
mode power = 28103.075
dip power = 28934.711
coupling efficiency = 0.971
Iteration = (45 / 100)
beta = 9.0
J = 9.2947e-01
grad_norm = 1.2422e-01
penalty = 0.441
mode power = 35342.692
dip power = 36301.386
coupling efficiency = 0.974
Iteration = (46 / 100)
beta = 10.0
J = 9.1725e-01
grad_norm = 2.0425e-01
penalty = 0.402
mode power = 22519.909
dip power = 23521.792
coupling efficiency = 0.957
Iteration = (47 / 100)
beta = 10.0
J = 9.3295e-01
grad_norm = 8.7255e-02
penalty = 0.397
mode power = 27879.178
dip power = 28662.293
coupling efficiency = 0.973
Iteration = (48 / 100)
beta = 10.0
J = 9.2987e-01
grad_norm = 1.6063e-01
penalty = 0.393
mode power = 36241.082
dip power = 37392.982
coupling efficiency = 0.969
Iteration = (49 / 100)
beta = 10.0
J = 9.3010e-01
grad_norm = 1.5227e-01
penalty = 0.388
mode power = 34755.844
dip power = 35869.977
coupling efficiency = 0.969
Iteration = (50 / 100)
beta = 10.0
J = 9.3480e-01
grad_norm = 8.6082e-02
penalty = 0.383
mode power = 24952.972
dip power = 25642.153
coupling efficiency = 0.973
Iteration = (51 / 100)
beta = 11.0
J = 9.2062e-01
grad_norm = 2.6132e-01
penalty = 0.353
mode power = 14742.606
dip power = 15421.940
coupling efficiency = 0.956
Iteration = (52 / 100)
beta = 11.0
J = 9.3850e-01
grad_norm = 6.0557e-02
penalty = 0.351
mode power = 30944.033
dip power = 31783.785
coupling efficiency = 0.974
Iteration = (53 / 100)
beta = 11.0
J = 9.2657e-01
grad_norm = 4.5544e-01
penalty = 0.349
mode power = 62661.433
dip power = 65175.091
coupling efficiency = 0.961
Iteration = (54 / 100)
beta = 11.0
J = 9.3726e-01
grad_norm = 1.3739e-01
penalty = 0.344
mode power = 26083.433
dip power = 26843.055
coupling efficiency = 0.972
Iteration = (55 / 100)
beta = 11.0
J = 9.3110e-01
grad_norm = 2.7552e-01
penalty = 0.340
mode power = 14073.962
dip power = 14582.365
coupling efficiency = 0.965
Iteration = (56 / 100)
beta = 12.0
J = 9.3442e-01
grad_norm = 2.0789e-01
penalty = 0.318
mode power = 10793.581
dip power = 11171.070
coupling efficiency = 0.966
Iteration = (57 / 100)
beta = 12.0
J = 9.3674e-01
grad_norm = 2.0010e-01
penalty = 0.316
mode power = 24160.809
dip power = 24951.189
coupling efficiency = 0.968
Iteration = (58 / 100)
beta = 12.0
J = 9.3609e-01
grad_norm = 4.0679e-01
penalty = 0.314
mode power = 70509.692
dip power = 72879.455
coupling efficiency = 0.967
Iteration = (59 / 100)
beta = 12.0
J = 9.3791e-01
grad_norm = 2.0113e-01
penalty = 0.311
mode power = 41804.340
dip power = 43143.020
coupling efficiency = 0.969
Iteration = (60 / 100)
beta = 12.0
J = 9.4025e-01
grad_norm = 2.5264e-01
penalty = 0.307
mode power = 25748.521
dip power = 26517.867
coupling efficiency = 0.971
Iteration = (61 / 100)
beta = 13.0
J = 9.4298e-01
grad_norm = 1.9770e-01
penalty = 0.289
mode power = 18771.299
dip power = 19314.514
coupling efficiency = 0.972
Iteration = (62 / 100)
beta = 13.0
J = 9.3831e-01
grad_norm = 2.4655e-01
penalty = 0.286
mode power = 36051.354
dip power = 37283.141
coupling efficiency = 0.967
Iteration = (63 / 100)
beta = 13.0
J = 9.4770e-01
grad_norm = 3.0916e-01
penalty = 0.284
mode power = 73330.091
dip power = 75124.741
coupling efficiency = 0.976
Iteration = (64 / 100)
beta = 13.0
J = 9.4116e-01
grad_norm = 2.3562e-01
penalty = 0.281
mode power = 48064.009
dip power = 49586.843
coupling efficiency = 0.969
Iteration = (65 / 100)
beta = 13.0
J = 9.4345e-01
grad_norm = 2.5314e-01
penalty = 0.279
mode power = 34732.187
dip power = 35758.010
coupling efficiency = 0.971
Iteration = (66 / 100)
beta = 14.0
J = 9.5072e-01
grad_norm = 1.4767e-01
penalty = 0.264
mode power = 28125.287
dip power = 28783.696
coupling efficiency = 0.977
Iteration = (67 / 100)
beta = 14.0
J = 9.4194e-01
grad_norm = 2.4715e-01
penalty = 0.262
mode power = 46559.660
dip power = 48091.621
coupling efficiency = 0.968
Iteration = (68 / 100)
beta = 14.0
J = 9.4917e-01
grad_norm = 2.2277e-01
penalty = 0.260
mode power = 72396.961
dip power = 74239.709
coupling efficiency = 0.975
Iteration = (69 / 100)
beta = 14.0
J = 9.4340e-01
grad_norm = 2.4728e-01
penalty = 0.258
mode power = 50687.983
dip power = 52298.049
coupling efficiency = 0.969
Iteration = (70 / 100)
beta = 14.0
J = 9.4637e-01
grad_norm = 2.1213e-01
penalty = 0.256
mode power = 46727.818
dip power = 48073.883
coupling efficiency = 0.972
Iteration = (71 / 100)
beta = 15.0
J = 9.4793e-01
grad_norm = 2.0206e-01
penalty = 0.245
mode power = 43807.749
dip power = 45051.273
coupling efficiency = 0.972
Iteration = (72 / 100)
beta = 15.0
J = 9.4802e-01
grad_norm = 2.3269e-01
penalty = 0.243
mode power = 65613.204
dip power = 67481.891
coupling efficiency = 0.972
Iteration = (73 / 100)
beta = 15.0
J = 9.4520e-01
grad_norm = 2.0955e-01
penalty = 0.241
mode power = 58289.876
dip power = 60136.353
coupling efficiency = 0.969
Iteration = (74 / 100)
beta = 15.0
J = 9.4843e-01
grad_norm = 1.9381e-01
penalty = 0.239
mode power = 59574.966
dip power = 61270.094
coupling efficiency = 0.972
Iteration = (75 / 100)
beta = 15.0
J = 9.4856e-01
grad_norm = 1.8808e-01
penalty = 0.237
mode power = 71769.585
dip power = 73817.117
coupling efficiency = 0.972
Iteration = (76 / 100)
beta = 16.0
J = 9.4920e-01
grad_norm = 2.2412e-01
penalty = 0.227
mode power = 34792.815
dip power = 35798.471
coupling efficiency = 0.972
Iteration = (77 / 100)
beta = 16.0
J = 9.5233e-01
grad_norm = 1.1844e-01
penalty = 0.225
mode power = 49022.100
dip power = 50286.898
coupling efficiency = 0.975
Iteration = (78 / 100)
beta = 16.0
J = 9.4676e-01
grad_norm = 4.5962e-01
penalty = 0.223
mode power = 112223.050
dip power = 115803.635
coupling efficiency = 0.969
Iteration = (79 / 100)
beta = 16.0
J = 9.5146e-01
grad_norm = 2.6840e-01
penalty = 0.222
mode power = 24794.677
dip power = 25465.193
coupling efficiency = 0.974
Iteration = (80 / 100)
beta = 16.0
J = 9.4763e-01
grad_norm = 2.8706e-01
penalty = 0.221
mode power = 20580.707
dip power = 21223.601
coupling efficiency = 0.970
Iteration = (81 / 100)
beta = 17.0
J = 9.4846e-01
grad_norm = 2.7118e-01
penalty = 0.213
mode power = 26519.913
dip power = 27347.167
coupling efficiency = 0.970
Iteration = (82 / 100)
beta = 17.0
J = 9.4705e-01
grad_norm = 2.4277e-01
penalty = 0.211
mode power = 83580.160
dip power = 86326.033
coupling efficiency = 0.968
Iteration = (83 / 100)
beta = 17.0
J = 9.4165e-01
grad_norm = 4.2922e-01
penalty = 0.210
mode power = 107209.997
dip power = 111366.379
coupling efficiency = 0.963
Iteration = (84 / 100)
beta = 17.0
J = 9.5012e-01
grad_norm = 2.7870e-01
penalty = 0.210
mode power = 26200.887
dip power = 26981.020
coupling efficiency = 0.971
Iteration = (85 / 100)
beta = 17.0
J = 9.4240e-01
grad_norm = 3.2850e-01
penalty = 0.209
mode power = 22240.250
dip power = 23088.709
coupling efficiency = 0.963
Iteration = (86 / 100)
beta = 18.0
J = 9.4756e-01
grad_norm = 2.9047e-01
penalty = 0.202
mode power = 21835.585
dip power = 22562.989
coupling efficiency = 0.968
Iteration = (87 / 100)
beta = 18.0
J = 9.4655e-01
grad_norm = 2.8581e-01
penalty = 0.201
mode power = 44635.553
dip power = 46177.909
coupling efficiency = 0.967
Iteration = (88 / 100)
beta = 18.0
J = 9.4568e-01
grad_norm = 1.4660e+00
penalty = 0.198
mode power = 191151.713
dip power = 197978.584
coupling efficiency = 0.966
Iteration = (89 / 100)
beta = 18.0
J = 9.0565e-01
grad_norm = 7.1548e-01
penalty = 0.200
mode power = 7787.385
dip power = 8413.092
coupling efficiency = 0.926
Iteration = (90 / 100)
beta = 18.0
J = 9.1608e-01
grad_norm = 5.4755e-01
penalty = 0.200
mode power = 3829.103
dip power = 4090.553
coupling efficiency = 0.936
Iteration = (91 / 100)
beta = 19.0
J = 9.1000e-01
grad_norm = 4.0796e-01
penalty = 0.196
mode power = 3154.019
dip power = 3393.009
coupling efficiency = 0.930
Iteration = (92 / 100)
beta = 19.0
J = 8.9532e-01
grad_norm = 4.8212e-01
penalty = 0.196
mode power = 3168.027
dip power = 3462.783
coupling efficiency = 0.915
Iteration = (93 / 100)
beta = 19.0
J = 9.1045e-01
grad_norm = 3.6817e-01
penalty = 0.196
mode power = 3353.528
dip power = 3605.939
coupling efficiency = 0.930
Iteration = (94 / 100)
beta = 19.0
J = 9.3083e-01
grad_norm = 2.9490e-01
penalty = 0.195
mode power = 3787.969
dip power = 3985.947
coupling efficiency = 0.950
Iteration = (95 / 100)
beta = 19.0
J = 9.4441e-01
grad_norm = 2.1215e-01
penalty = 0.194
mode power = 4620.248
dip power = 4793.893
coupling efficiency = 0.964
Iteration = (96 / 100)
beta = 20.0
J = 9.3738e-01
grad_norm = 2.9252e-01
penalty = 0.188
mode power = 5972.530
dip power = 6246.091
coupling efficiency = 0.956
Iteration = (97 / 100)
beta = 20.0
J = 9.4350e-01
grad_norm = 2.2733e-01
penalty = 0.186
mode power = 9200.513
dip power = 9562.867
coupling efficiency = 0.962
Iteration = (98 / 100)
beta = 20.0
J = 9.5092e-01
grad_norm = 1.1442e-01
penalty = 0.184
mode power = 15828.774
dip power = 16330.019
coupling efficiency = 0.969
Iteration = (99 / 100)
beta = 20.0
J = 9.4448e-01
grad_norm = 1.1112e-01
penalty = 0.182
mode power = 30118.514
dip power = 31287.115
coupling efficiency = 0.963
Iteration = (100 / 100)
beta = 20.0
J = 9.3400e-01
grad_norm = 1.4951e-01
penalty = 0.179
mode power = 60020.849
dip power = 63053.450
coupling efficiency = 0.952
Ultimately, we get all the information to assess the optimization results.
[18]:
obj_vals = np.array(history_dict["values"])
ce_vals = np.array(history_dict["coupl_eff"])
pen_vals = np.array(history_dict["penalty"])
final_par_density = history_dict["params"][-1]
final_beta = history_dict["beta"][-1]
[19]:
# just to inspect design at different iterations
def unfold_params(params):
params = np.concatenate((np.fliplr(np.copy(params)), params), axis=1)
return params
params1 = history_dict["params"][32]
params1_full = pre_process(params1, beta=final_beta)
params1_full = include_constant_regions(
params1_full, circ_center=[qe_pos.center[0], qe_pos.center[1]], circ_radius=non_etch_r
)
params1_full = unfold_params(params1_full)
params2 = history_dict["params"][-1]
params2_full = pre_process(params2, beta=final_beta)
params2_full = include_constant_regions(
params2_full, circ_center=[qe_pos.center[0], qe_pos.center[1]], circ_radius=non_etch_r
)
params2_full = unfold_params(params2_full)
[20]:
plt.imshow(
1 - np.flipud(params1_full.T),
cmap="gray",
vmin=0,
vmax=1,
extent=[wg_length, cr_l + wg_length, -cr_w / 2, cr_w / 2],
)
plt.plot(dp_source.center[0], 0, "r*")
plt.show()
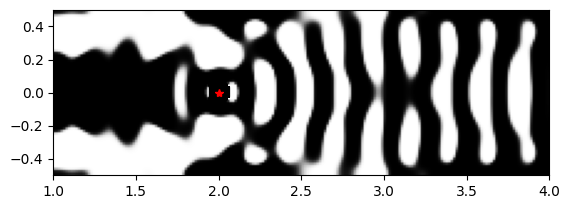
[21]:
plt.imshow(
1 - np.flipud(params2_full.T),
cmap="gray",
vmin=0,
vmax=1,
extent=[wg_length, cr_l + wg_length, -cr_w / 2, cr_w / 2],
)
plt.plot(dp_source.center[0], 0, "r*")
plt.show()
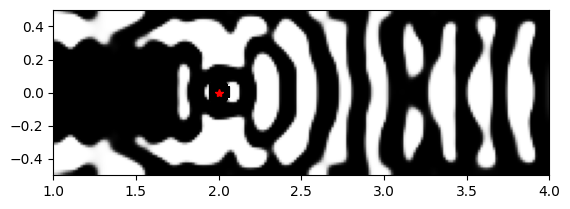
Results#
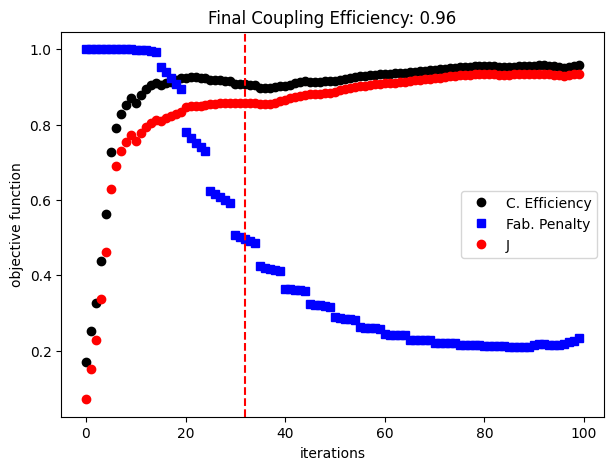
The following figure shows how coupling efficiency and the fabrication penalty have evolved along the optimization process. The coupling efficiency quickly rises above 0.8, and along the binarization process, we can observe two large drops before a more stable final optimization stage. The formation of resonant modes sensitive to the small structural changes can potentially explain this behavior. The discontinuities in the fabrication penalty curve are caused by the increments in the projection parameter beta at each 5 iterations.
[22]:
fig, ax = plt.subplots(1, 1, figsize=(7, 5))
ax.plot(ce_vals, "ko", label="C. Efficiency")
ax.plot(pen_vals, "bs", label="Fab. Penalty")
ax.plot(history_dict["values"], "ro", label="J")
ax.set_xlabel("iterations")
ax.set_ylabel("objective function")
ax.set_title(f"Final Coupling Efficiency: {ce_vals[-1]:.2f}")
ax.axvline(x=32, color="r", linestyle="--")
ax.legend()
plt.show()
Interestingly, fully binarizing the design from iteration 32 produced a device with great coupling efficiency (~97%), but minimal purcell enhancement
[23]:
plt.plot([np.linalg.norm(grad) for grad in history_dict["gradients"]])
plt.ylabel("norm(grad)")
plt.xlabel("iteration")
[23]:
Text(0.5, 0, 'iteration')
Makes a nice animation of the design parameters and gradient evolution during the optimization
[24]:
import matplotlib.animation as animation
from mpl_toolkits.axes_grid1 import make_axes_locatable
fig, axs = plt.subplots(nrows=2, ncols=1)
gradients = history_dict["gradients"]
params = history_dict["params"]
gradients = [unfold_params(grad).T for grad in gradients]
params = [unfold_params(init_par).T] + [unfold_params(1.0 - p).T for p in params]
div = make_axes_locatable(axs[1])
div0 = make_axes_locatable(axs[0])
cax = div.append_axes("top", size="5%", pad=0.05)
cax0 = div0.append_axes("bottom", size="5%", pad=0.05)
cax0.axis("off")
def animate(i):
im_g = axs[1].imshow(
gradients[i], interpolation="none", vmin=np.min(gradients[i]), vmax=np.max(gradients[i])
)
axs[0].imshow(params[i], interpolation="none", cmap="gray", vmin=0, vmax=1)
axs[1].axis("off")
axs[0].axis("off")
cax.cla()
fig.colorbar(im_g, cax=cax, orientation="horizontal").ax.xaxis.set_ticks_position("top")
axs[0].set_title(f"iteration {i}")
anim = animation.FuncAnimation(fig, animate, frames=100, blit=False, interval=500)
anim.save("autograd_anim.mp4", fps=2.0)
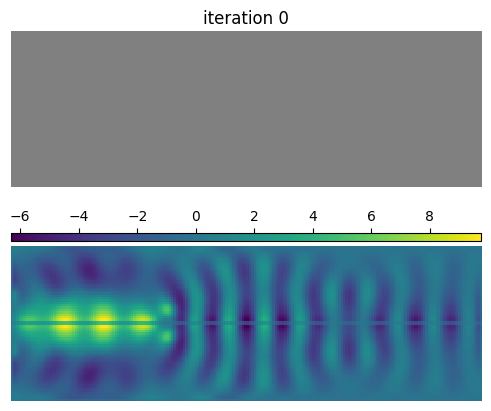
Interestingly, the final quantum emitter light extractor resembles a nanocavity, even though we have considered only the coupling efficiency into the output waveguide in the optimization. We have DBR mirrors on both sides of the dipole. However, on the left side, the mirror has only a few periods and partially reflects the radiation, which couples to the output waveguide.
[25]:
# here, removing substrate improved performance, but also blue-shifted cavity resonance a bit.
fig, ax = plt.subplots(1, figsize=(10, 4))
# Substrate layer.
substrate = td.Structure(
geometry=td.Box.from_bounds(
rmin=(-eff_inf, -eff_inf, -eff_inf), rmax=(eff_inf, eff_inf, -wg_thick / 2)
),
medium=td.Medium(permittivity=1.0),
)
sim_final = make_adjoint_sim(params2, beta=final_beta, unfold=True)
sim_final = sim_final.updated_copy(monitors=[field_monitor_xy, mode_monitor] + field_monitor)
sim_final.plot_eps(
z=0,
source_alpha=0,
monitor_alpha=0,
ax=ax,
)
plt.show()
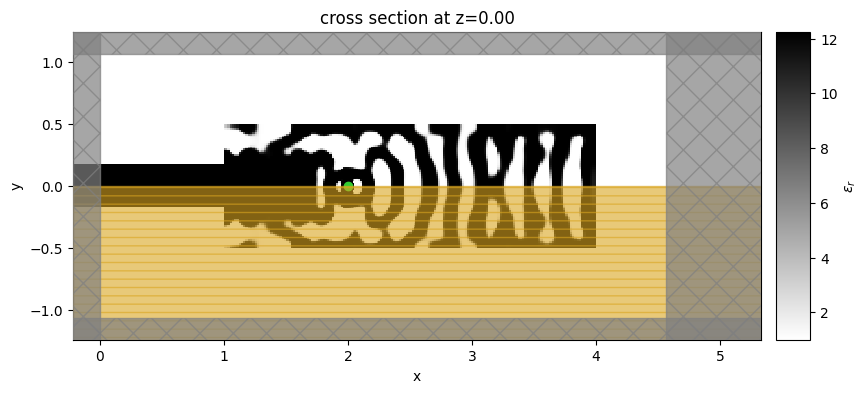
To better understand the resultant design, let’s simulate the final structure to obtain its spectral response and field distribution.
[26]:
sim_data_final = web.run(sim_final, task_name="final QE light extractor")
18:56:12 CEST Created task 'final QE light extractor' with task_id 'fdve-7b5a7f2b-9dda-46bc-8519-0af1e82273c8' and task_type 'FDTD'.
View task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-7b5a7f2b-9d da-46bc-8519-0af1e82273c8'.
Task folder: 'default'.
18:56:14 CEST Maximum FlexCredit cost: 0.058. Minimum cost depends on task execution details. Use 'web.real_cost(task_id)' to get the billed FlexCredit cost after a simulation run.
18:56:15 CEST status = queued
To cancel the simulation, use 'web.abort(task_id)' or 'web.delete(task_id)' or abort/delete the task in the web UI. Terminating the Python script will not stop the job running on the cloud.
18:56:27 CEST starting up solver
running solver
18:56:36 CEST early shutoff detected at 24%, exiting.
status = postprocess
18:56:41 CEST status = success
18:56:43 CEST View simulation result at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-7b5a7f2b-9d da-46bc-8519-0af1e82273c8'.
18:56:47 CEST loading simulation from simulation_data.hdf5
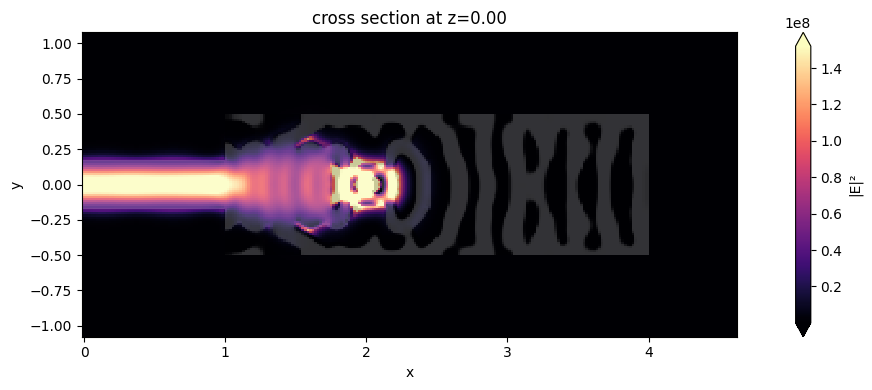
In this cavity-like system, the extraction efficiency of photons from the QE into the collection waveguide mode is proportional to \(\beta\times C_{wg}\), where the \(\beta\)-factor quantifies the fraction of the QE spontaneous emission emitted in the cavity mode, and \(C_{wg}\) is the fraction of the cavity photons coupled to the guided mode
A. Enderlin, Y. Ota, R. Ohta, N. Kumagai, S. Ishida, S. Iwamoto, and Y. Arakawa, "High guided mode–cavity mode coupling for an efficient extraction of spontaneous emission of a single quantum dot embedded in a photonic crystal nanobeam cavity," Phys. Rev. B 86, 075314 (2012) DOI: 10.1103/PhysRevB.86.075314. By the field distribution image below, we can see a cavity mode resonance, which should increase the Purcell factor at the QE
position, thus contributing to a higher \(\beta\)-factor. At the same time, the partial reflection mirror at the left side was potentially optimized to adjust \(C_{wg}\).
[27]:
f, ax1 = plt.subplots(1, 1, figsize=(12, 4), tight_layout=True)
sim_data_final.plot_field("field_xy", "E", "abs^2", z=0, ax=ax1, f=freqs[0])
plt.show()
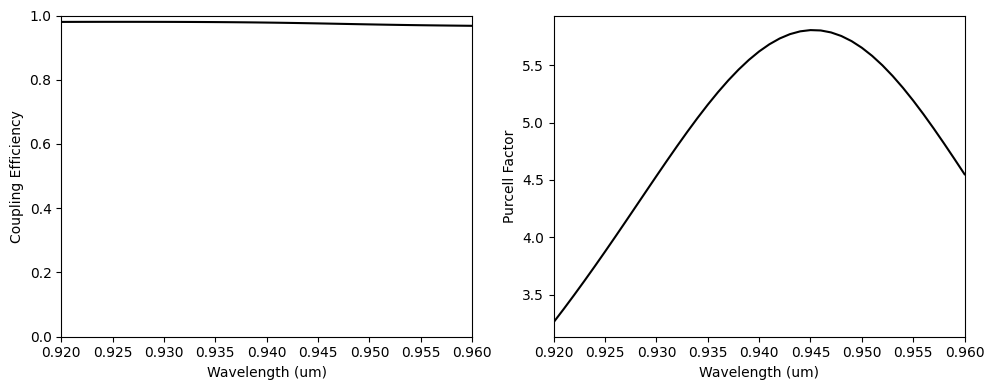
To conclude, we will calculate the final coupling efficiency and the cavity Purcell value. The coupling efficiency is above 80% along an extensive wavelength range, and we have confirmed the Purcell enhancement.
[28]:
# Coupling efficiency.
mode_amps = sim_data_final["mode_monitor"].amps.sel(direction="-", mode_index=0)
mode_power = np.abs(mode_amps) ** 2
dip_power = np.zeros(n_wl)
for i in range(len(field_monitor)):
field_mon = sim_data_final[f"field_monitor_{i}"]
dip_power += np.abs(field_mon.flux)
coup_eff = mode_power / dip_power
# Purcell factor.
bulk_power = ((2 * np.pi * freqs) ** 2 / (12 * np.pi)) * (td.MU_0 * n_wg / td.C_0)
bulk_power = bulk_power * 2 ** (2 * np.sum(np.abs(sim_final.symmetry)))
purcell = dip_power / bulk_power
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4), tight_layout=True)
ax1.plot(wl_range, coup_eff, "-k")
ax1.set_xlabel("Wavelength (um)")
ax1.set_ylabel("Coupling Efficiency")
ax1.set_ylim(0, 1)
ax1.set_xlim(wl - bw / 2, wl + bw / 2)
ax2.plot(wl_range, purcell, "-k")
ax2.set_xlabel("Wavelength (um)")
ax2.set_ylabel("Purcell Factor")
ax2.set_xlim(wl - bw / 2, wl + bw / 2)
plt.show()
[29]:
print(np.max(coup_eff.values))
print(np.min(coup_eff.values))
0.9685581762335904
0.8529465434212032
Export to GDS#
The Simulation object has the .to_gds_file convenience function to export the final design to a GDS file. In addition to a file name, it is necessary to set a cross-sectional plane (z = 0 in this case) on which to evaluate the geometry, a frequency to evaluate the permittivity, and a permittivity_threshold to define the shape boundaries in custom
mediums. See the GDS export notebook for a detailed example on using .to_gds_file and other GDS related functions.
[30]:
# make the misc/ directory to store the GDS file if it doesn't exist already
import os
if not os.path.exists("./misc/"):
os.mkdir("./misc/")
sim_final.to_gds_file(
fname="./misc/inv_des_light_extractor_autograd.gds",
z=0,
permittivity_threshold=(eps_max + eps_min) / 2,
frequency=freq,
)